Development of antisense RNA-mediated quantifiable inhibition for metabolic regulation
- PMID: 33717978
- PMCID: PMC7921874
- DOI: 10.1016/j.mec.2021.e00168
Development of antisense RNA-mediated quantifiable inhibition for metabolic regulation
Abstract
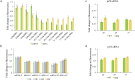
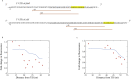
Trans-regulating elements such as noncoding RNAs are crucial in modifying cells, and has shown broad application in synthetic biology, metabolic engineering and RNA therapies. Although effective, titration of the regulatory levels of such elements is less explored. Encouraged by the need of fine-tuning cellular functions, we studied key parameters of the antisense RNA design including oligonucleotide length, targeting region and relative dosage to achieve differentiated inhibition. We determined a 30-nucleotide configuration that renders efficient and robust inhibition. We found that by targeting the core RBS region proportionally, quantifiable inhibition levels can be rationally obtained. A mathematic model was established accordingly with refined energy terms and successfully validated by depicting the inhibition levels for genomic targets. Additionally, we applied this fine-tuning approach for 4-hydroxycoumarin biosynthesis by simultaneous and quantifiable knockdown of multiple targets, resulting in a 3.58-fold increase in titer of the engineered strain comparing to that of the non-regulated. We believe the developed tool is broadly compatible and provides an extra layer of control in modifying living systems.
Keywords: 4-Hydroxycoumarin; Antisense RNA; Core RBS region; Fine-tuning; Quantifiable inhibition.
© 2021 The Authors.
Conflict of interest statement
The authors claim no conflict of interest.
Figures




References
-
- Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. - PubMed
-
- Bailey J.E. Toward a science of metabolic engineering. Science. 1991;252:1668–1675. - PubMed
-
- Bremer H., Dennis P.P. American Society for Microbiology. Chapter, Modulation of chemical composition and other parameters of the cell by growth rate; Washington (DC): 1996. Escherichia coli and Salmonella: Cellular and Molecular Biology; pp. 1553–1569.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

