FOXA2-Interacting FOXP2 Prevents Epithelial-Mesenchymal Transition of Breast Cancer Cells by Stimulating E-Cadherin and PHF2 Transcription
- PMID: 33718155
- PMCID: PMC7947682
- DOI: 10.3389/fonc.2021.605025
FOXA2-Interacting FOXP2 Prevents Epithelial-Mesenchymal Transition of Breast Cancer Cells by Stimulating E-Cadherin and PHF2 Transcription
Abstract
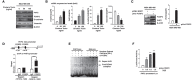
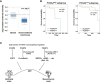
FOXP2, a member of forkhead box transcription factor family, was first identified as a language-related gene that played an important role in language learning and facial movement. In addition, FOXP2 was also suggested regulating the progression of cancer cells. In previous studies, we found that FOXA2 inhibited epithelial-mesenchymal transition (EMT) in breast cancer cells. In this study, by identifying FOXA2-interacting proteins from FOXA2-pull-down cell lysates with Mass Spectrometry Analysis, we found that FOXP2 interacted with FOXA2. After confirming the interaction between FOXP2 and FOXA2 through Co-IP and immunofluorescence assays, we showed a correlated expression of FOXP2 and FOXA2 existing in clinical breast cancer samples. The overexpression of FOXP2 attenuated the mesenchymal phenotype whereas the stable knockdown of FOXP2 promoted EMT in breast cancer cells. Even though FOXP2 was believed to act as a transcriptional repressor in most cases, we found that FOXP2 could activate the expression of tumor suppressor PHF2. Meanwhile, we also found that FOXP2 could endogenously bind to the promoter of E-cadherin and activate its transcription. This transcriptional activity of FOXP2 relied on its interaction with FOXA2. Furthermore, the stable knockdown of FOXP2 enhanced the metastatic capacity of breast cancer cells in vivo. Together, the results suggested that FOXP2 could inhibit EMT by activating the transcription of certain genes, such as E-cadherin and PHF2, in concert with FOXA2 in breast cancer cells.
Keywords: E-cadherin; FOXA2 transcription factor; FOXP2 transcription factor; PHF2; epithelial-mesenchymal transition of breast cancer.
Copyright © 2021 Liu, Chen, Guo, Li, Zhang, Tan, Yu and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Clarke M, Collins R, Darby S, Davies C, Elphinstone P, Evans V, et al. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15-year survival: an overview of the randomised trials. Lancet (2005) 366:2087–106. 10.1016/S0140-6736(05)67887-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

