Mining of RNA Methylation-Related Genes and Elucidation of Their Molecular Biology in Gallbladder Carcinoma
- PMID: 33718182
- PMCID: PMC7947712
- DOI: 10.3389/fonc.2021.621806
Mining of RNA Methylation-Related Genes and Elucidation of Their Molecular Biology in Gallbladder Carcinoma
Abstract
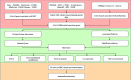
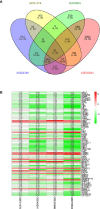
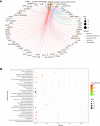
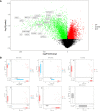
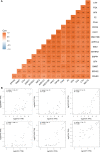
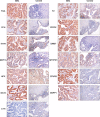
Gallbladder carcinoma (GBC), which has high invasion and metastasis risks, remains the most common biliary tract malignancy. Surgical resection for GBC is the only effective treatment, but most patients miss the opportunity for curative surgery because of a lack of timely diagnosis. The aim of this study was to identify and verify early candidate diagnostic and prognostic RNA methylation related genes for GBC via integrated transcriptome bioinformatics analysis. Lists of GBC-related genes and methylation-related genes were collected from public databases to screen differentially expressed genes (DEGs) by using the limma package and the RobustRankAggreg (RRA) package. The core genes were collected with batch effects corrected by the RRA algorithm through protein interaction network analysis, signaling pathway enrichment analysis and gene ranking. Four modules obtained from four public microarray datasets were found to be related to GBC, and FGA, F2, HAO1, CFH, PIPOX, ITIH4, GNMT, MAT1A, MTHFD1, HPX, CTH, EPHX2, HSD17B6, AKR1C4, CFHR3, ENNP1, and NAT2 were revealed to be potential hub genes involved in methylation-related pathways and bile metabolism-related pathways. Among these, FGA, CFH, F2, HPX, and PIPOX were predicted to be methylated genes in GBC, but POPIX had no modification sites for RNA methylation. Furthermore, survival analysis of TCGA (the Cancer Genome Atlas) database showed that six genes among the hub genes, FGA, CFH, ENPP1, CFHR3, ITIH4, and NAT2, were highly expressed and significantly correlated with worse prognosis. Gene correlation analysis revealed that the FGA was positively correlated with the ENPP1, NAT2, and CFHR3, while CFH was positively correlated with the NAT2, CFHR3, and FGA. In addition, the results of immunohistochemistry (IHC) showed that the expressions of FGA, F2, CFH, PIPOX, ITIH4, GNMT, MAT1A, MTHFD1, HPX, CFHR3, NAT2, and ENPP1 were higher in GBC tissues than that in control tissues. In conclusion, two genes, FGA and CFH, were identified as RNA methylation-related genes also involved in bile metabolism in GBC, which may be novel biomarkers to early diagnose and evaluate prognosis for GBC.
Keywords: RNA methylation; bile metabolism; bioinformatics; biomarkers; differentially expressed genes; gallbladder carcinoma.
Copyright © 2021 Yang, Chen, Yu, Luo, Li, Zhou and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Unravelling of the comparative Transcriptomic Profile of Gallbladder Cancer using mRNA sequencing.Mol Biol Rep. 2022 Jul;49(7):6395-6403. doi: 10.1007/s11033-022-07448-4. Epub 2022 Apr 25. Mol Biol Rep. 2022. PMID: 35469389
-
Identification of key DNA methylation-driven genes in prostate adenocarcinoma: an integrative analysis of TCGA methylation data.J Transl Med. 2019 Sep 18;17(1):311. doi: 10.1186/s12967-019-2065-2. J Transl Med. 2019. PMID: 31533842 Free PMC article.
-
RNA sequencing reveals differentially expressed genes as potential diagnostic and prognostic indicators of gallbladder carcinoma.Oncotarget. 2015 Aug 21;6(24):20661-71. doi: 10.18632/oncotarget.3861. Oncotarget. 2015. PMID: 25970782 Free PMC article.
-
A systematic review with in silico analysis on transcriptomic profile of gallbladder carcinoma.Semin Oncol. 2020 Dec;47(6):398-408. doi: 10.1053/j.seminoncol.2020.02.012. Epub 2020 Oct 24. Semin Oncol. 2020. PMID: 33162112
-
Epigenetic changes in carcinogenesis of gallbladder.Indian J Surg Oncol. 2013 Dec;4(4):356-61. doi: 10.1007/s13193-013-0240-0. Epub 2013 Apr 4. Indian J Surg Oncol. 2013. PMID: 24426757 Free PMC article. Review.
Cited by
-
A novel hypoxia-driven gene signature that can predict the prognosis of hepatocellular carcinoma.Bioengineered. 2022 May;13(5):12193-12210. doi: 10.1080/21655979.2022.2073943. Bioengineered. 2022. PMID: 35549979 Free PMC article.
-
Potential Molecular Markers for Cholecystic Inflammation-Induced Carcinogenesis Based on RNA-Seq Gene Screening.Comb Chem High Throughput Screen. 2025;28(6):931-943. doi: 10.2174/0113862073287686240409082130. Comb Chem High Throughput Screen. 2025. PMID: 38716548
-
Complement system-related genes in stomach adenocarcinoma: Prognostic signature, immune landscape, and drug resistance.Front Genet. 2022 Sep 8;13:903421. doi: 10.3389/fgene.2022.903421. eCollection 2022. Front Genet. 2022. PMID: 36159981 Free PMC article.
-
The miR-590-3p/CFHR3/STAT3 signaling pathway promotes cell proliferation and metastasis in hepatocellular carcinoma.Aging (Albany NY). 2022 Jul 18;14(14):5783-5799. doi: 10.18632/aging.204178. Epub 2022 Jul 18. Aging (Albany NY). 2022. PMID: 35852862 Free PMC article.
-
Biliary Tract Cancers: Treatment Updates and Future Directions in the Era of Precision Medicine and Immuno-Oncology.Front Oncol. 2021 Nov 15;11:768009. doi: 10.3389/fonc.2021.768009. eCollection 2021. Front Oncol. 2021. PMID: 34868996 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

