Comprehensive Analysis of Expression Regulation for RNA m6A Regulators With Clinical Significance in Human Cancers
- PMID: 33718187
- PMCID: PMC7946859
- DOI: 10.3389/fonc.2021.624395
Comprehensive Analysis of Expression Regulation for RNA m6A Regulators With Clinical Significance in Human Cancers
Abstract
Background: N6-methyladenosine (m6A), the most abundant chemical modification on eukaryotic messenger RNA (mRNA), is modulated by three class of regulators namely "writers," "erasers," and "readers." Increasing studies have shown that aberrant expression of m6A regulators plays broad roles in tumorigenesis and progression. However, it is largely unknown regarding the expression regulation for RNA m6A regulators in human cancers.
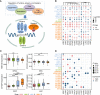
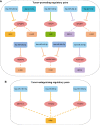
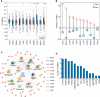
Results: Here we characterized the expression profiles of RNA m6A regulators in 13 cancer types with The Cancer Genome Atlas (TCGA) data. We showed that METTL14, FTO, and ALKBH5 were down-regulated in most cancers, whereas YTHDF1 and IGF2BP3 were up-regulated in 12 cancer types except for thyroid carcinoma (THCA). Survival analysis further revealed that low expression of several m6A regulators displayed longer overall survival times. Then, we analyzed microRNA (miRNA)-regulated and DNA methylation-regulated expression changes of m6A regulators in pan-cancer. In total, we identified 158 miRNAs and 58 DNA methylation probes (DMPs) involved in expression regulation for RNA m6A regulators. Furthermore, we assessed the survival significance of those regulatory pairs. Among them, 10 miRNAs and 7 DMPs may promote cancer initiation and progression; conversely, 3 miRNA/mRNA pairs in kidney renal clear cell carcinoma (KIRC) may exert tumor-suppressor function. These findings are indicative of their potential prognostic values. Finally, we validated two of those miRNA/mRNA pairs (hsa-miR-1307-3p/METTL14 and hsa-miR-204-5p/IGF2BP3) that could serve a critical role for potential clinical application in KIRC patients.
Conclusions: Our findings highlighted the importance of upstream regulation (miRNA and DNA methylation) governing m6A regulators' expression in pan-cancer. As a result, we identified several informative regulatory pairs for prognostic stratification. Thus, our study provides new insights into molecular mechanisms of m6A modification in human cancers.
Keywords: DNA methylation; N6-methyladenosine; The Cancer Genome Atlas; microRNA; prognosis.
Copyright © 2021 Liu, Wang, Teng, Zhang and Song.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Pan-Cancer Prognostic, Immunity, Stemness, and Anticancer Drug Sensitivity Characterization of N6-Methyladenosine RNA Modification Regulators in Human Cancers.Front Mol Biosci. 2021 Jun 4;8:644620. doi: 10.3389/fmolb.2021.644620. eCollection 2021. Front Mol Biosci. 2021. PMID: 34150845 Free PMC article.
-
Analysis and validation of m6A regulatory network: a novel circBACH2/has-miR-944/HNRNPC axis in breast cancer progression.J Transl Med. 2021 Dec 24;19(1):527. doi: 10.1186/s12967-021-03196-4. J Transl Med. 2021. PMID: 34952600 Free PMC article.
-
Contributions and Prognostic Values of N6-Methyladenosine RNA Methylation Regulators in Hepatocellular Carcinoma.Front Genet. 2021 Jan 15;11:614566. doi: 10.3389/fgene.2020.614566. eCollection 2020. Front Genet. 2021. PMID: 33519919 Free PMC article.
-
The potential role of RNA N6-methyladenosine in Cancer progression.Mol Cancer. 2020 May 12;19(1):88. doi: 10.1186/s12943-020-01204-7. Mol Cancer. 2020. PMID: 32398132 Free PMC article. Review.
-
Crosstalk Between m6A RNA Methylation and miRNA Biogenesis in Cancer: An Unholy Nexus.Mol Biotechnol. 2024 Nov;66(11):3042-3058. doi: 10.1007/s12033-023-00921-w. Epub 2023 Oct 13. Mol Biotechnol. 2024. PMID: 37831403 Review.
Cited by
-
Potential prognosis index for m6A-related mRNA in cholangiocarcinoma.BMC Cancer. 2022 Jun 7;22(1):620. doi: 10.1186/s12885-022-09665-3. BMC Cancer. 2022. PMID: 35672673 Free PMC article.
-
Construction and validation of an m6A RNA methylation regulator prognostic model for early-stage clear cell renal cell carcinoma.Oncol Lett. 2022 Jun 10;24(2):250. doi: 10.3892/ol.2022.13370. eCollection 2022 Aug. Oncol Lett. 2022. PMID: 35761938 Free PMC article.
-
Multi-omics analysis of N6-methyladenosine reader IGF2BP3 as a promising biomarker in pan-cancer.Front Immunol. 2023 Jan 25;14:1071675. doi: 10.3389/fimmu.2023.1071675. eCollection 2023. Front Immunol. 2023. PMID: 36761737 Free PMC article.
-
N6-Methyladenosine RNA Modification: An Emerging Immunotherapeutic Approach to Turning Up Cold Tumors.Front Cell Dev Biol. 2021 Sep 20;9:736298. doi: 10.3389/fcell.2021.736298. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34616742 Free PMC article. Review.
-
The emerging roles of N6-methyladenosine RNA modifications in thyroid cancer.Eur J Med Res. 2023 Nov 1;28(1):475. doi: 10.1186/s40001-023-01382-2. Eur J Med Res. 2023. PMID: 37915103 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

