Fecal Multidimensional Assay for Non-Invasive Detection of Colorectal Cancer: Fecal Immunochemical Test, Stool DNA Mutation, Methylation, and Intestinal Bacteria Analysis
- PMID: 33718241
- PMCID: PMC7947614
- DOI: 10.3389/fonc.2021.643136
Fecal Multidimensional Assay for Non-Invasive Detection of Colorectal Cancer: Fecal Immunochemical Test, Stool DNA Mutation, Methylation, and Intestinal Bacteria Analysis
Abstract
Background: Fecal immunochemical test (FIT), DNA mutation, DNA methylation, and microbial dysbiosis all showed promising in colorectal cancer (CRC) non-invasive detection. We assessed CRC detection with an assay combining all these strategies and investigated the effect of clinical features on the performance of this comprehensive test.
Methods: We performed a multidimensional analysis study using stool samples collected from 108 patients with CRC, 18 patients with colorectal adenoma, and 36 individuals with no evidence of colorectal disease. The multidimensional analysis of stool samples including FIT, stool DNA (sDNA) tests for three methylated genes (Septin9, NDRG4, BMP3) and three mutated genes (KRAS, BRAF, PI3KCA) using next generation sequencing as well as detection of stool bacteria level of Fusobacterium nucleatum and Parvimonas micra using qPCR method. We used a linear support vector classification model to analyze the data.
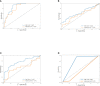
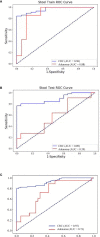
Results: The sensitivity of FIT alone was 69.4% for CRC and 11.1% for adenoma. Separately, the sensitivity of the detection of intestinal bacteria, DNA mutation, and DNA methylation for CRC was 58.3, 50.0, and 51.9%, respectively. The combination of FIT and sDNA tests had a sensitivity of 81.5% for CRC (AUC: 0.93, better than FIT alone, P = 0.017) and 27.8% for adenoma with 94.4% specificity. Sensitivity of the multidimensional test to detect CRC with stage II (84.6%) and III (91.9%) CRC was relatively higher (88.2%) than that of patients with stage I (60.0%) and stage IV (75.0%) (P = 0.024). The rate of CRC detection increased with tumor size (P = 0.008) and age (P = 0.04). Interestingly, the rate of CRC detection was higher in smoking persons than non-smokers with marginal significance (P = 0.08).
Conclusions: The multidimensional assay of stool samples combining FIT and stool DNA tests further improved the diagnostic sensitivity for CRC. This could provide new approach for improvement of CRC screening and further demonstrations are warranted.
Keywords: cancer screening; colorectal cancer; fecal biomarker; human gut microbiome; methylation.
Copyright © 2021 Mo, Wang, Han, Xiang, Dai, Zhao, Pei, Su, Ma, Li, Wang, Cai, Wang, Liu and Cai.
Conflict of interest statement
HuiW, PZ, FP, ZS, CM and RL were employed by company Singlera Genomics (Shanghai). The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Feasibility of quantification based on novel evaluation with stool DNA and fecal immunochemical test for colorectal cancer detection.BMC Gastroenterol. 2022 Aug 13;22(1):384. doi: 10.1186/s12876-022-02470-z. BMC Gastroenterol. 2022. PMID: 35963995 Free PMC article.
-
Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia.Clin Gastroenterol Hepatol. 2013 Oct;11(10):1313-8. doi: 10.1016/j.cgh.2013.04.023. Epub 2013 Apr 29. Clin Gastroenterol Hepatol. 2013. PMID: 23639600
-
A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma.Mutat Res Rev Mutat Res. 2019 Jan-Mar;779:45-57. doi: 10.1016/j.mrrev.2019.01.003. Epub 2019 Jan 16. Mutat Res Rev Mutat Res. 2019. PMID: 31097151
-
[The clinical value of multi-target stool fecal immunochemical test-DNA in early screening and diagnosis for colorectal cancer].Zhonghua Yi Xue Za Zhi. 2022 Sep 6;102(33):2607-2613. doi: 10.3760/cma.j.cn112137-20220430-00974. Zhonghua Yi Xue Za Zhi. 2022. PMID: 36058686 Chinese.
-
Noninvasive fecal testing for colorectal cancer.Clin Chim Acta. 2022 Jan 1;524:123-131. doi: 10.1016/j.cca.2021.10.030. Epub 2021 Oct 28. Clin Chim Acta. 2022. PMID: 34756863 Review.
Cited by
-
Combined DNA Methylation and Gastric Microbiome Marker Predicts Helicobacter pylori-Negative Gastric Cancer.Gut Liver. 2024 Jul 15;18(4):611-620. doi: 10.5009/gnl230348. Epub 2024 Mar 21. Gut Liver. 2024. PMID: 38509701 Free PMC article.
-
A Comparison of Single and Combined Schemes of Asia-Pacific Colorectal Screening, Faecal Immunochemical and Stool Deoxyribonucleic Acid Testing for Community Colorectal Cancer Screening.J Multidiscip Healthc. 2023 Mar 1;16:571-586. doi: 10.2147/JMDH.S398997. eCollection 2023. J Multidiscip Healthc. 2023. PMID: 36883167 Free PMC article.
-
The global research of microbiota in colorectal cancer screening: a bibliometric and visualization analysis.Front Oncol. 2023 May 5;13:1169369. doi: 10.3389/fonc.2023.1169369. eCollection 2023. Front Oncol. 2023. PMID: 37213286 Free PMC article.
-
Developing a Nomogram for Predicting Colorectal Cancer and Its Precancerous Lesions Based on Data from Three Non-Invasive Screening Tools, APCS, FIT, and sDNA.J Multidiscip Healthc. 2024 Jun 14;17:2891-2901. doi: 10.2147/JMDH.S465286. eCollection 2024. J Multidiscip Healthc. 2024. PMID: 38903878 Free PMC article.
-
Novel multiplex stool-based assay for the detection of early-stage colon cancer in a Chinese population.World J Gastroenterol. 2022 Jun 28;28(24):2705-2732. doi: 10.3748/wjg.v28.i24.2705. World J Gastroenterol. 2022. PMID: 35979157 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

