Specific and Sensitive Diagnosis of BCOR-ITD in Various Cancers by Digital PCR
- PMID: 33718245
- PMCID: PMC7948083
- DOI: 10.3389/fonc.2021.645512
Specific and Sensitive Diagnosis of BCOR-ITD in Various Cancers by Digital PCR
Abstract
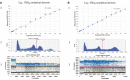
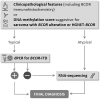
BCOR is an epigenetic regulator altered by various mechanisms including BCOR-internal tandem duplication (BCOR-ITD) in a wide range of cancers. Six different BCOR-ITD in the 3'-part of the coding sequence of exon 15 have been reported ranging from 89 to 114 bp in length. BCOR-ITD is a common genetic alteration found in clear cell sarcoma of the kidney and primitive myxoid mesenchymal tumor of infancy (PMMTI) and it characterizes a new type of central nervous system tumor: "CNS tumor with BCOR-ITD". It can also be detected in undifferentiated round cell sarcoma (URCS) and in high-grade endometrial stromal sarcoma (HGESS). Therefore, it is of utmost importance to search for this genetic alteration in these cancers with the most frequent technique being RNA-sequencing. Here, we developed a new droplet PCR assay (dPCR) to detect the six sequences characterizing BCOR-ITD. To achieve this goal, we used a single colored probe to detect both the duplicated region and the normal sequence that acts as a reference. We first generated seven synthetic DNA sequences: ITD0 (the normal sequence) and ITD1 to ITD6 (the duplicated sequences described in the literature) and then we set up the optima dPCR conditions. We validated our assay on 19 samples from a representative panel of human tumors (9 HGNET-BCOR, 5 URCS, 3 HGESS, and 2 PMMTI) in which BCOR-ITD status was known using at least one other method including RNA sequencing, RT-PCR or DNA-methylation profiling for CNS tumors. Our results showed that our technique was 100% sensitive and specific. DPCR detected BCOR-ITD in 13/19 of the cases; in the remaining 6 cases additional RNA-sequencing revealed BCOR gene fusions. To conclude, in the era of histomolecular classification of human tumors, our modified dPCR assay is of particular interest to detect BCOR-ITD since it is a robust and less expensive test that can be applied to a broad spectrum of cancers that share this alteration.
Keywords: BCOR-internal tandem duplication; FFPE tissue; HGNET-BCOR; diagnostic marker; digital PCR assay.
Copyright © 2021 Barets, Appay, Heinisch, Battistella, Bouvier, Chotard, Le Loarer, Macagno, Perbet, Pissaloux, Rousseau, Tauziède-Espariat, Varlet, Vasiljevic, Colin, Fina and Figarella-Branger.
Conflict of interest statement
FF is Scientific Director of ID-Solutions, Grabels, France. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

