Proteomic Analyses of Acinetobacter baumannii Clinical Isolates to Identify Drug Resistant Mechanism
- PMID: 33718272
- PMCID: PMC7943614
- DOI: 10.3389/fcimb.2021.625430
Proteomic Analyses of Acinetobacter baumannii Clinical Isolates to Identify Drug Resistant Mechanism
Abstract
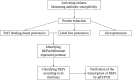
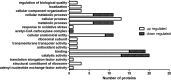
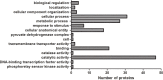
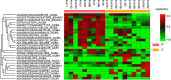
Acinetobacter baumannii is one of the main causes of nosocomial infections. Increasing numbers of multidrug-resistant Acinetobacter baumannii cases have been reported in recent years, but its antibiotic resistance mechanism remains unclear. We studied 9 multidrug-resistant (MDR) and 10 drug-susceptible Acinetobacter baumannii clinical isolates using Label free, TMT labeling approach and glycoproteomics analysis to identify proteins related to drug resistance. Our results showed that 164 proteins exhibited different expressions between MDR and drug-susceptible isolates. These differential proteins can be classified into six groups: a. proteins related to antibiotic resistance, b. membrane proteins, membrane transporters and proteins related to membrane formation, c. Stress response-related proteins, d. proteins related to gene expression and protein translation, e. metabolism-related proteins, f. proteins with unknown function or other functions containing biofilm formation and virulence. In addition, we verified seven proteins at the transcription level in eight clinical isolates by using quantitative RT-PCR. Results showed that four of the selected proteins have positive correlations with the protein level. This study provided an insight into the mechanism of antibiotic resistance of multidrug-resistant Acinetobacter baumannii.
Keywords: Acinetobacter baumannii; TMT; antibiotic resistance; glycopeptides; label free; proteomic.
Copyright © 2021 Wang, Li, Wang, Yang, Zou and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Virulence properties of multidrug resistant ocular isolates of Acinetobacter baumannii.Curr Eye Res. 2014 Jul;39(7):695-704. doi: 10.3109/02713683.2013.873055. Epub 2014 Feb 6. Curr Eye Res. 2014. PMID: 24502411
-
Phenotypic and molecular characterization of Acinetobacter baumannii isolates causing lower respiratory infections among ICU patients.Microb Pathog. 2019 Mar;128:75-81. doi: 10.1016/j.micpath.2018.12.023. Epub 2018 Dec 15. Microb Pathog. 2019. PMID: 30562602
-
Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: analysis of genotype, antimicrobial susceptibility, and biofilm formation.Ann Clin Microbiol Antimicrob. 2018 Jun 1;17(1):24. doi: 10.1186/s12941-018-0277-6. Ann Clin Microbiol Antimicrob. 2018. PMID: 29859115 Free PMC article.
-
Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy.Expert Rev Anti Infect Ther. 2010 Jan;8(1):71-93. doi: 10.1586/eri.09.108. Expert Rev Anti Infect Ther. 2010. PMID: 20014903 Review.
-
Insights Into Mechanisms of Biofilm Formation in Acinetobacter baumannii and Implications for Uropathogenesis.Front Cell Infect Microbiol. 2020 May 29;10:253. doi: 10.3389/fcimb.2020.00253. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32547965 Free PMC article. Review.
Cited by
-
Deciphering Antibiotic-Targeted Metabolic Pathways in Acinetobacter baumannii: Insights from Transcriptomics and Genome-Scale Metabolic Modeling.Life (Basel). 2024 Sep 2;14(9):1102. doi: 10.3390/life14091102. Life (Basel). 2024. PMID: 39337886 Free PMC article.
-
Deciphering the Intracellular Action of the Antimicrobial Peptide A11 via an In-Depth Analysis of Its Effect on the Global Proteome of Acinetobacter baumannii.ACS Infect Dis. 2024 Aug 9;10(8):2795-2813. doi: 10.1021/acsinfecdis.4c00160. Epub 2024 Jul 29. ACS Infect Dis. 2024. PMID: 39075773 Free PMC article.
-
Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii.Biomolecules. 2022 Jun 30;12(7):922. doi: 10.3390/biom12070922. Biomolecules. 2022. PMID: 35883478 Free PMC article.
-
Antibiotic Resistance Diagnosis in ESKAPE Pathogens-A Review on Proteomic Perspective.Diagnostics (Basel). 2023 Mar 7;13(6):1014. doi: 10.3390/diagnostics13061014. Diagnostics (Basel). 2023. PMID: 36980322 Free PMC article. Review.
-
A novel strategy to characterize the pattern of β-lactam antibiotic-induced drug resistance in Acinetobacter baumannii.Sci Rep. 2023 Jun 6;13(1):9177. doi: 10.1038/s41598-023-36475-9. Sci Rep. 2023. PMID: 37280269 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

