Rv3722c Promotes Mycobacterium tuberculosis Survival in Macrophages by Interacting With TRAF3
- PMID: 33718275
- PMCID: PMC7947218
- DOI: 10.3389/fcimb.2021.627798
Rv3722c Promotes Mycobacterium tuberculosis Survival in Macrophages by Interacting With TRAF3
Abstract
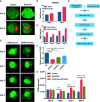
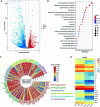
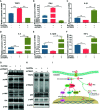
Mycobacterium tuberculosis (M.tb) secretes numerous proteins to interfere with host immune response for its long-term survival. As one of the top abundant M.tb secreted proteins, Rv3722c was found to be essential for bacilli growth. However, it remains elusive how this protein interferes with the host immune response and regulates M.tb survival. Here, we confirmed that Rv3722c interacted with host TRAF3 to promote M.tb replication in macrophages. Knock-down of TRAF3 attenuated the effect of Rv3722c on the intracellular M.tb survival. The interaction between Rv3722c and TRAF3 hampered MAPK and NF-κB pathways, resulting in a significant increase of IFN-β expression and decrease of IL-1β, IL-6, IL-12p40, and TNF-α expression. Our study revealed that Rv3722c interacted with TRAF3 and interrupted its downstream pathways to promote M.tb survival in macrophages. These findings facilitate further understanding of the mechanism of M.tb secreted proteins in regulating the host cell immune response and promoting its intracellular survival.
Keywords: Mycobacterium tuberculosis; Rv3722c; TRAF3; cytokines; intracellular survival.
Copyright © 2021 Lei, Cao, Xu, Yang, Xu, Zhou, Dong, Wu, Rahman, Tyagi, Zhao, Chen and Cao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bramucci E., Milano T., Pascarella S. (2011). Genomic distribution and heterogeneity of MocR-like transcriptional factors containing a domain belonging to the superfamily of the pyridoxal-5’-phosphate dependent enzymes of fold type I. Biochem. Biophys. Res. Commun. 415 (1), 88–93. 10.1016/j.bbrc.2011.10.017 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

