Development of a Rapid and Fully Automated Multiplex Real-Time PCR Assay for Identification and Differentiation of Vibrio cholerae and Vibrio parahaemolyticus on the BD MAX Platform
- PMID: 33718286
- PMCID: PMC7947656
- DOI: 10.3389/fcimb.2021.639473
Development of a Rapid and Fully Automated Multiplex Real-Time PCR Assay for Identification and Differentiation of Vibrio cholerae and Vibrio parahaemolyticus on the BD MAX Platform
Abstract
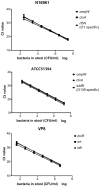
Vibrio cholerae and Vibrio parahaemolyticus are common diarrheal pathogens of great public health concern. A multiplex TaqMan-based real-time PCR assay was developed on the BD MAX platform; this assay can simultaneously detect and differentiate V. cholerae and V. parahaemolyticus directly from human fecal specimens. The assay includes two reactions. One reaction, BDM-VC, targets the gene ompW, the cholera toxin (CT) coding gene ctxA, the O1 serogroup specific gene rfbN, and the O139 serogroup specific gene wbfR of V. cholerae. The other, BDM-VP, targets the gene toxR and the toxin coding genes tdh and trh of V. parahaemolyticus. In addition, each reaction contains a sample process control. When evaluated with spiked stool samples, the detection limit of the BD MAX assay was 195-780 CFU/ml for V. cholerae and 46-184 CFU/ml for V. parahaemolyticus, and the amplification efficiency of all genes was between 95 and 115%. The assay showed 100% analytical specificity, using 63 isolates. The BD MAX assay was evaluated for its performance compared with conventional real-time PCR after automated DNA extraction steps, using 164 retrospective stool samples. The overall percent agreement between the BD MAX assay and real-time PCR was ≥ 98.8%; the positive percent agreement was 85.7% for ompW, 100% for toxR/tdh, and lower (66.7%) for trh because of a false negative. This is the first report to evaluate the usage of the BD MAX open system in detection and differentiation of V. cholerae and V. parahaemolyticus directly from human samples.
Keywords: BD MAX; Vibrio cholerae; Vibrio parahaemolyticus; multiplex; real-time PCR.
Copyright © 2021 Li, Guan, Wang, Gao, Feng, Li, Diao, Zhao, Kan and Zhang.
Conflict of interest statement
A PCT patent application has been filed in China. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Bonnin-Jusserand M., Copin S., Le Bris C., Brauge T., Gay M., Brisabois A., et al. . (2019). Vibrio species involved in seafood-borne outbreaks (Vibrio cholerae, V. parahaemolyticus and V. vulnificus): review of microbiological versus recent molecular detection methods in seafood products. Crit. Rev. Food Sci. Nutr. 59 (4), 597–610. 10.1080/10408398.2017.1384715 - DOI - PubMed
-
- Harrington S. M., Buchan B. W., Doern C., Fader R., Ferraro M. J., Pillai D. R., et al. . (2015). Multicenter evaluation of the BD max enteric bacterial panel PCR assay for rapid detection of Salmonella spp., Shigella spp., Campylobacter spp. (C. jejuni and C. coli), and Shiga toxin 1 and 2 genes. J. Clin. Microbiol. 53 (5), 1639–1647. 10.1128/JCM.03480-14 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases