Conformational Changes of Glutamine 5'-Phosphoribosylpyrophosphate Amidotransferase for Two Substrates Analogue Binding: Insight from Conventional Molecular Dynamics and Accelerated Molecular Dynamics Simulations
- PMID: 33718330
- PMCID: PMC7953260
- DOI: 10.3389/fchem.2021.640994
Conformational Changes of Glutamine 5'-Phosphoribosylpyrophosphate Amidotransferase for Two Substrates Analogue Binding: Insight from Conventional Molecular Dynamics and Accelerated Molecular Dynamics Simulations
Abstract
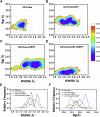
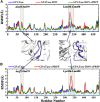
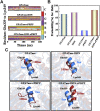
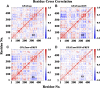
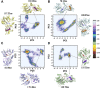
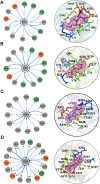
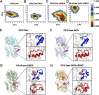
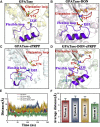
Glutamine 5'-phosphoribosylpyrophosphate amidotransferase (GPATase) catalyzes the synthesis of phosphoribosylamine, pyrophosphate, and glutamate from phosphoribosylpyrophosphate, as well as glutamine at two sites (i.e., glutaminase and phosphoribosylpyrophosphate sites), through a 20 Å NH3 channel. In this study, conventional molecular dynamics (cMD) simulations and enhanced sampling accelerated molecular dynamics (aMD) simulations were integrated to characterize the mechanism for coordination catalysis at two separate active sites in the enzyme. Results of cMD simulations illustrated the mechanism by which two substrate analogues, namely, DON and cPRPP, affect the structural stability of GPATase from the perspective of dynamic behavior. aMD simulations obtained several key findings. First, a comparison of protein conformational changes in the complexes of GPATase-DON and GPATase-DON-cPRPP showed that binding cPRPP to the PRTase flexible loop (K326 to L350) substantially effected the formation of the R73-DON salt bridge. Moreover, only the PRTase flexible loop in the GPATase-DON-cPRPP complex could remain closed and had sufficient space for cPRPP binding, indicating that binding of DON to the glutamine loop had an impact on the PRTase flexible loop. Finally, both DON and cPRPP tightly bonded to the two domains, thereby inducing the glutamine loop and the PRTase flexible loop to move close to each other. This movement facilitated the transfer of NH3 via the NH3 channel. These theoretical results are useful to the ongoing research on efficient inhibitors related to GPATase.
Keywords: 5′-phosphoribosylpyrophosphate amidotransferase; accelerated molecular dynamics simulations; conformational changes; molecular dynamics simulations; substrates analogue.
Copyright © 2021 Li, Chen, Huang, Zhang, Yuan, Chang, Li and Han.
Conflict of interest statement
Author HC was employed by the company Jilin Province TeyiFood Biotechnology Company Limited. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling Editor declared a shared affiliation, though no other collaboration, with several of the authors (CL, SC, TH, FZ, JY, WL, and WH).
Figures



References
-
- Berendsen H. J. C., Postma J. P. M., Van Gunsteren W. F., Dinola A., Haak J. R. (1998). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81 (8), 3684–3690. 10.1063/1.448118 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

