Minimize the Xylitol Production in Saccharomyces cerevisiae by Balancing the Xylose Redox Metabolic Pathway
- PMID: 33718341
- PMCID: PMC7953151
- DOI: 10.3389/fbioe.2021.639595
Minimize the Xylitol Production in Saccharomyces cerevisiae by Balancing the Xylose Redox Metabolic Pathway
Abstract
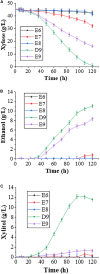
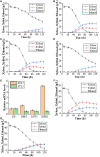
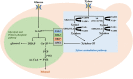
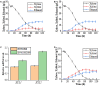
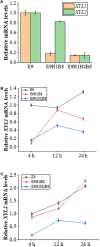
Xylose is the second most abundant sugar in lignocellulose, but it cannot be used as carbon source by budding yeast Saccharomyces cerevisiae. Rational promoter elements engineering approaches were taken for efficient xylose fermentation in budding yeast. Among promoters surveyed, HXT7 exhibited the best performance. The HXT7 promoter is suppressed in the presence of glucose and derepressed by xylose, making it a promising candidate to drive xylose metabolism. However, simple ectopic expression of both key xylose metabolic genes XYL1 and XYL2 by the HXT7 promoter resulted in massive accumulation of the xylose metabolic byproduct xylitol. Through the HXT7-driven expression of a reported redox variant, XYL1-K270R, along with optimized expression of XYL2 and the downstream pentose phosphate pathway genes, a balanced xylose metabolism toward ethanol formation was achieved. Fermented in a culture medium containing 50 g/L xylose as the sole carbon source, xylose is nearly consumed, with less than 3 g/L xylitol, and more than 16 g/L ethanol production. Hence, the combination of an inducible promoter and redox balance of the xylose utilization pathway is an attractive approach to optimizing fuel production from lignocellulose.
Keywords: Saccharomyces cerevisiae; expression; promoter; xylitol; xylose.
Copyright © 2021 Zhu, Zhang, Zhu, Jia, Li, Xiao and Cao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Baghel R. S., Trivedi N., Gupta V., Neori A., Reddy C. R. K., Lali A., et al. (2015). Biorefining of marine macroalgal biomass for production of biofuel and commodity chemicals. Green Chem. 17 2436–2443. 10.1039/c4gc02532f - DOI
-
- Bao X., Gao D., Qu Y., Wang Z., Walfridssion M., Hahnhagerbal B. (1997). Effect on product formation in recombinant Saccharomyces cerevisiae strains expressing different levels of xylose metabolic genes. Chin. J. Biotechnol. 13:225. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

