Alternatively Splicing Interactomes Identify Novel Isoform-Specific Partners for NSD2
- PMID: 33718354
- PMCID: PMC7947288
- DOI: 10.3389/fcell.2021.612019
Alternatively Splicing Interactomes Identify Novel Isoform-Specific Partners for NSD2
Abstract
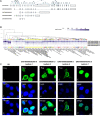
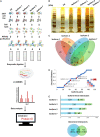
Nuclear receptor SET domain protein (NSD2) plays a fundamental role in the pathogenesis of Wolf-Hirschhorn Syndrome (WHS) and is overexpressed in multiple human myelomas, but its protein-protein interaction (PPI) patterns, particularly at the isoform/exon levels, are poorly understood. We explored the subcellular localizations of four representative NSD2 transcripts with immunofluorescence microscopy. Next, we used label-free quantification to perform immunoprecipitation mass spectrometry (IP-MS) analyses of the transcripts. Using the interaction partners for each transcript detected in the IP-MS results, we identified 890 isoform-specific PPI partners (83% are novel). These PPI networks were further divided into four categories of the exon-specific interactome. In these exon-specific PPI partners, two genes, RPL10 and HSPA8, were successfully confirmed by co-immunoprecipitation and Western blotting. RPL10 primarily interacted with Isoforms 1, 3, and 5, and HSPA8 interacted with all four isoforms, respectively. Using our extended NSD2 protein interactions, we constructed an isoform-level PPI landscape for NSD2 to serve as reference interactome data for NSD2 spliceosome-level studies. Furthermore, the RNA splicing processes supported by these isoform partners shed light on the diverse roles NSD2 plays in WHS and myeloma development. We also validated the interactions using Western blotting, RPL10, and the three NSD2 (Isoform 1, 3, and 5). Our results expand gene-level NSD2 PPI networks and provide a basis for the treatment of NSD2-related developmental diseases.
Keywords: NSD2; RPL10; alternatively splicing; isoform; protein–protein interaction.
Copyright © 2021 Wang, Chen, Zhao, Chen, Song, Li and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
From Wolf-Hirschhorn syndrome to NSD2 haploinsufficiency: a shifting paradigm through the description of a new case and a review of the literature.Ital J Pediatr. 2022 May 12;48(1):72. doi: 10.1186/s13052-022-01267-w. Ital J Pediatr. 2022. PMID: 35550183 Free PMC article. Review.
-
The first familial NSD2 cases with a novel variant in a Chinese father and daughter with atypical WHS facial features and a 7.5-year follow-up of growth hormone therapy.BMC Med Genomics. 2020 Dec 4;13(1):181. doi: 10.1186/s12920-020-00831-9. BMC Med Genomics. 2020. PMID: 33276791 Free PMC article.
-
Loss of function in NSD2 causes DNA methylation signature similar to that in Wolf-Hirschhorn syndrome.Genet Med Open. 2024 Mar 14;2:101838. doi: 10.1016/j.gimo.2024.101838. eCollection 2024. Genet Med Open. 2024. PMID: 39669601 Free PMC article.
-
Case report: A de novo NSD2 truncating variant in a child with Rauch-Steindl syndrome.Front Pediatr. 2023 Jun 7;11:1064783. doi: 10.3389/fped.2023.1064783. eCollection 2023. Front Pediatr. 2023. PMID: 37351323 Free PMC article.
-
Drug Discovery Targeting Nuclear Receptor Binding SET Domain Protein 2 (NSD2).J Med Chem. 2023 Aug 24;66(16):10991-11026. doi: 10.1021/acs.jmedchem.3c00948. Epub 2023 Aug 14. J Med Chem. 2023. PMID: 37578463 Free PMC article. Review.
References
-
- Anderson-Schmidt H., Beltcheva O., Brandon M. D., Byrne E. M., Diehl E. J., Duncan L., et al. (2013). Selected rapporteur summaries from the XX world congress of psychiatric genetics, Hamburg, Germany, october 14-18, 2012. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 162 96–121. 10.1002/ajmg.b.32132 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

