Transcriptome Analysis of Dnmt3l Knock-Out Mice Derived Multipotent Mesenchymal Stem/Stromal Cells During Osteogenic Differentiation
- PMID: 33718357
- PMCID: PMC7947861
- DOI: 10.3389/fcell.2021.615098
Transcriptome Analysis of Dnmt3l Knock-Out Mice Derived Multipotent Mesenchymal Stem/Stromal Cells During Osteogenic Differentiation
Abstract
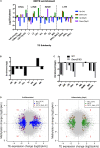
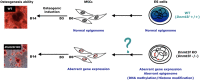
Multipotent mesenchymal stem/stromal cells (MSCs) exhibit great potential for cell-based therapy. Proper epigenomic signatures in MSCs are important for the maintenance and the subsequent differentiation potential. The DNA methyltransferase 3-like (DNMT3L) that was mainly expressed in the embryonic stem (ES) cells and the developing germ cells plays an important role in shaping the epigenetic landscape. Here, we report the reduced colony forming ability and impaired in vitro osteogenesis in Dnmt3l-knockout-mice-derived MSCs (Dnmt3l KO MSCs). By comparing the transcriptome between undifferentiated Dnmt3l KO MSCs and the MSCs from the wild-type littermates, some of the differentially regulated genes (DEGs) were found to be associated with bone-morphology-related phenotypes. On the third day of osteogenic induction, differentiating Dnmt3l KO MSCs were enriched for genes associated with nucleosome structure, peptide binding and extracellular matrix modulation. Differentially expressed transposable elements in many subfamilies reflected the change of corresponding regional epigenomic signatures. Interestingly, DNMT3L protein is not expressed in cultured MSCs. Therefore, the observed defects in Dnmt3l KO MSCs are unlikely a direct effect from missing DNMT3L in this cell type; instead, we hypothesized them as an outcome of the pre-deposited epigenetic signatures from the DNMT3L-expressing progenitors. We observed that 24 out of the 107 upregulated DEGs in Dnmt3l KO MSCs were hypermethylated in their gene bodies of DNMT3L knock-down ES cells. Among these 24 genes, some were associated with skeletal development or homeostasis. However, we did not observe reduced bone development, or reduced bone density through aging in vivo. The stronger phenotype in vitro suggested the involvement of potential spreading and amplification of the pre-deposited epigenetic defects over passages, and the contribution of oxidative stress during in vitro culture. We demonstrated that transient deficiency of epigenetic co-factor in ES cells or progenitor cells caused compromised property in differentiating cells much later. In order to facilitate safer practice in cell-based therapy, we suggest more in-depth examination shall be implemented for cells before transplantation, even on the epigenetic level, to avoid long-term risk afterward.
Keywords: DNA methylation; DNMT3L; bone-marrow MSCs; epigenetics; osteogenesis.
Copyright © 2021 Yang, Lu, Lee, Hsiao, Yen, Cheng, Hsu, Tsai, Chen, Liu, Chen and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

