Activity of the SNARE Protein SNAP29 at the Endoplasmic Reticulum and Golgi Apparatus
- PMID: 33718375
- PMCID: PMC7945952
- DOI: 10.3389/fcell.2021.637565
Activity of the SNARE Protein SNAP29 at the Endoplasmic Reticulum and Golgi Apparatus
Abstract
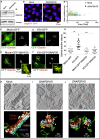
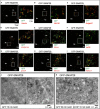
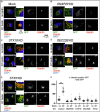
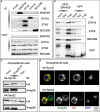
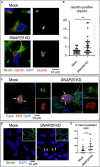
Snap29 is a conserved regulator of membrane fusion essential to complete autophagy and to support other cellular processes, including cell division. In humans, inactivating SNAP29 mutations causes CEDNIK syndrome, a rare multi-systemic disorder characterized by congenital neuro-cutaneous alterations. The fibroblasts of CEDNIK patients show alterations of the Golgi apparatus (GA). However, whether and how Snap29 acts at the GA is unclear. Here we investigate SNAP29 function at the GA and endoplasmic reticulum (ER). As part of the elongated structures in proximity to these membrane compartments, a pool of SNAP29 forms a complex with Syntaxin18, or with Syntaxin5, which we find is required to engage SEC22B-loaded vesicles. Consistent with this, in HeLa cells, in neuroepithelial stem cells, and in vivo, decreased SNAP29 activity alters GA architecture and reduces ER to GA trafficking. Our data reveal a new regulatory function of Snap29 in promoting secretory trafficking.
Keywords: Golgi apparatus; SEC22B; SNAP29 gene; SNARE protein; Syntaxin 5; endoplasmic reticulum; vesicle fusion.
Copyright © 2021 Morelli, Speranza, Pellegrino, Beznoussenko, Carminati, Garré, Mironov, Onorati and Vaccari.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

