M2 macrophage-derived exosomal microRNA-155-5p promotes the immune escape of colon cancer by downregulating ZC3H12B
- PMID: 33718596
- PMCID: PMC7932913
- DOI: 10.1016/j.omto.2021.02.005
M2 macrophage-derived exosomal microRNA-155-5p promotes the immune escape of colon cancer by downregulating ZC3H12B
Abstract
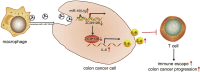
Previous evidence has highlighted M2 macrophage regulation of cancer cells via exosome shuttling of microRNAs (miRNAs or miRs). The current study set out to explore the possible role of M2 macrophage-derived exosomal miR-155-5p in regard to immune escape of colon cancer cells. Experimental data from quantitative reverse-transcriptase PCR (qRT-PCR) and western blot analysis revealed highly expressed miR-155-5p and interleukin (IL)-6 and poorly expressed ZC3H12B in M2 macrophage-derived exosomes. Additionally, miR-155-5p could be transferred by M2 macrophage-isolated exosomes to colon cancer cells, which targeted ZC3H12B by binding to the 3¢ UTR, as identified by dual luciferase reporter gene. Meanwhile, gain- and loss-of function experimentation on miR-155-5p and ZC3H12B in SW48 and HT29 cells cocultured with M2 macrophage-secreted exosomes demonstrated that miR-155-5p overexpression or ZC3H12B silencing promoted the proliferation and antiapoptosis ability of SW48 and HT29 cells, as well as augmenting the CD3+ T cell proliferation and the proportion of interferon (IFN)-γ+ T cells. Xenograft models confirmed that M2 macrophage-derived exosomal miR-155-5p reduced the ZC3H12B expression to upregulate IL-6, which consequently induced immune escape and tumor formation. Collectively, our findings indicated that M2 macrophage-derived exosomal miR-155-5p can potentially promote the immune escape of colon cancer by impairing ZC3H12B-mediated IL-6 stability reduction, thereby promoting the occurrence and development of colon cancer.
Keywords: M2 macrophages; ZC3H12B; colon cancer; exosomes; immune escape; microRNA-155-5p.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Exosomal miR-183-5p Shuttled by M2 Polarized Tumor-Associated Macrophage Promotes the Development of Colon Cancer via Targeting THEM4 Mediated PI3K/AKT and NF-κB Pathways.Front Oncol. 2021 Jun 25;11:672684. doi: 10.3389/fonc.2021.672684. eCollection 2021. Front Oncol. 2021. PMID: 34249713 Free PMC article.
-
M2 macrophage-derived exosomal miR-145-5p protects against the hypoxia/reoxygenation-induced pyroptosis of cardiomyocytes by inhibiting TLR4 expression.Ann Transl Med. 2022 Dec;10(24):1376. doi: 10.21037/atm-22-6109. Ann Transl Med. 2022. PMID: 36660616 Free PMC article.
-
Colon Cancer-Derived Exosomal LncRNA-XIST Promotes M2-like Macrophage Polarization by Regulating PDGFRA.Int J Mol Sci. 2024 Oct 24;25(21):11433. doi: 10.3390/ijms252111433. Int J Mol Sci. 2024. PMID: 39518984 Free PMC article.
-
M2 macrophage-derived exosomal miR-1911-5p promotes cell migration and invasion in lung adenocarcinoma by down-regulating CELF2 -activated ZBTB4 expression.Anticancer Drugs. 2023 Feb 1;34(2):238-247. doi: 10.1097/CAD.0000000000001414. Epub 2023 Nov 18. Anticancer Drugs. 2023. PMID: 36730375
-
M2 macrophage-derived exosomal miR-486-5p influences the differentiation potential of bone marrow mesenchymal stem cells and osteoporosis.Aging (Albany NY). 2023 Sep 25;15(18):9499-9520. doi: 10.18632/aging.205031. Epub 2023 Sep 25. Aging (Albany NY). 2023. PMID: 37751585 Free PMC article.
Cited by
-
Tryptophan metabolism and disposition in cancer biology and immunotherapy.Biosci Rep. 2022 Nov 30;42(11):BSR20221682. doi: 10.1042/BSR20221682. Biosci Rep. 2022. PMID: 36286592 Free PMC article.
-
Exosome derived from tumor-associated macrophages: biogenesis, functions, and therapeutic implications in human cancers.Biomark Res. 2023 Nov 19;11(1):100. doi: 10.1186/s40364-023-00538-w. Biomark Res. 2023. PMID: 37981718 Free PMC article. Review.
-
The homeostatic function of Regnase-2 restricts neuroinflammation.FASEB J. 2023 Mar;37(3):e22798. doi: 10.1096/fj.202201978R. FASEB J. 2023. PMID: 36753401 Free PMC article.
-
Exosomes and ferroptosis: roles in tumour regulation and new cancer therapies.PeerJ. 2022 Apr 26;10:e13238. doi: 10.7717/peerj.13238. eCollection 2022. PeerJ. 2022. PMID: 35497192 Free PMC article. Review.
-
Macrophage-Derived Small Extracellular Vesicles in Multiple Diseases: Biogenesis, Function, and Therapeutic Applications.Front Cell Dev Biol. 2022 Jun 27;10:913110. doi: 10.3389/fcell.2022.913110. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35832790 Free PMC article. Review.
References
-
- Chien C.W., Hou P.C., Wu H.C., Chang Y.L., Lin S.C., Lin S.C., Lin B.W., Lee J.C., Chang Y.J., Sun H.S., Tsai S.J. Targeting TYRO3 inhibits epithelial-mesenchymal transition and increases drug sensitivity in colon cancer. Oncogene. 2016;35:5872–5881. - PubMed
-
- Vermeer N.C.A., Claassen Y.H.M., Derks M.G.M., Iversen L.H., van Eycken E., Guren M.G., Mroczkowski P., Martling A., Johansson R., Vandendael T. Treatment and Survival of Patients with Colon Cancer Aged 80 Years and Older: A EURECCA International Comparison. Oncologist. 2018;23:982–990. - PMC - PubMed
-
- Gangireddy V.G.R., Coleman T., Kanneganti P., Talla S., Annapureddy A.R., Amin R., Parikh S. Polypectomy versus surgery in early colon cancer: size and location of colon cancer affect long-term survival. Int. J. Colorectal Dis. 2018;33:1349–1357. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

