Mimicking H3 Substrate Arginine in the Design of G9a Lysine Methyltransferase Inhibitors for Cancer Therapy: A Computational Study for Structure-Based Drug Design
- PMID: 33718701
- PMCID: PMC7948220
- DOI: 10.1021/acsomega.0c04710
Mimicking H3 Substrate Arginine in the Design of G9a Lysine Methyltransferase Inhibitors for Cancer Therapy: A Computational Study for Structure-Based Drug Design
Abstract
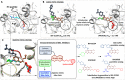
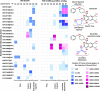
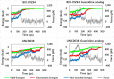
G9a protein methyltransferase is a potential epigenetic drug target in different cancers and other disease conditions overexpressing the enzyme. G9a is responsible for the H3K9 dimethylation mark, which epigenetically regulates gene expression. Arg8 and Lys9 of the H3 substrate peptide are the two crucial residues for substrate-specific recognition and methylation. Several substrate competitive inhibitors are reported for the potent inhibition of G9a by incorporating lysine mimic groups in the inhibitor design. In this study, we explored the concept of arginine mimic strategy. The hydrophobic segment of the reported inhibitors BIX-01294 and UNC0638 was replaced by a guanidine moiety (side-chain moiety of arginine). The newly substituted guanidine moieties of the inhibitors were positioned similar to the Arg8 of the substrate peptide in molecular docking. Additionally, improved reactivity of the guanidine-substituted inhibitors was observed in density functional theory studies. Molecular dynamics, molecular mechanics Poisson-Boltzmann surface area binding free energy, linear interaction energy, and potential mean force calculated from steered molecular dynamics simulations of the newly designed analogues show enhanced conformational stability and improved H-bond potential and binding affinity toward the target G9a. Moreover, the presence of both lysine and arginine mimics together shows a drastic increase in the binding affinity of the inhibitor towards G9a. Hence, we propose incorporating a guanidine group to imitate the substrate arginine's side chain in the inhibitor design to improve the potency of G9a inhibitors.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Delineating the active site architecture of G9a lysine methyltransferase through substrate and inhibitor binding mode analysis: a molecular dynamics study.J Biomol Struct Dyn. 2019 Jul;37(10):2581-2592. doi: 10.1080/07391102.2018.1491422. Epub 2018 Nov 17. J Biomol Struct Dyn. 2019. PMID: 30047835
-
Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.Nat Struct Mol Biol. 2009 Mar;16(3):312-7. doi: 10.1038/nsmb.1560. Epub 2009 Feb 15. Nat Struct Mol Biol. 2009. PMID: 19219047 Free PMC article.
-
Identification of novel quinoline inhibitor for EHMT2/G9a through virtual screening.Biochimie. 2020 Jan;168:220-230. doi: 10.1016/j.biochi.2019.11.006. Epub 2019 Nov 20. Biochimie. 2020. PMID: 31756401
-
Insights for the design of protein lysine methyltransferase G9a inhibitors.Future Med Chem. 2019 May;11(9):993-1014. doi: 10.4155/fmc-2018-0396. Epub 2019 May 29. Future Med Chem. 2019. PMID: 31141392 Review.
-
Recent progress in histone methyltransferase (G9a) inhibitors as anticancer agents.Eur J Med Chem. 2019 Oct 1;179:537-546. doi: 10.1016/j.ejmech.2019.06.072. Epub 2019 Jun 28. Eur J Med Chem. 2019. PMID: 31276898 Review.
Cited by
-
SETDB1 as a cancer target: challenges and perspectives in drug design.RSC Med Chem. 2024 Mar 19;15(5):1424-1451. doi: 10.1039/d3md00366c. eCollection 2024 May 22. RSC Med Chem. 2024. PMID: 38799223 Free PMC article. Review.
-
EHMT2/G9a as an Epigenetic Target in Pediatric and Adult Brain Tumors.Int J Mol Sci. 2021 Oct 19;22(20):11292. doi: 10.3390/ijms222011292. Int J Mol Sci. 2021. PMID: 34681949 Free PMC article. Review.
-
G9a an Epigenetic Therapeutic Strategy for Neurodegenerative Conditions: From Target Discovery to Clinical Trials.Med Res Rev. 2025 May;45(3):985-1015. doi: 10.1002/med.22096. Epub 2025 Jan 6. Med Res Rev. 2025. PMID: 39763018 Free PMC article. Review.
References
-
- Tachibana M.; Sugimoto K.; Fukushima T.; Shinkai Y. SET Domain-Containing Protein, G9a, Is a Novel Lysine-Preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. J. Biol. Chem. 2001, 276, 25309–25317. 10.1074/jbc.m101914200. - DOI - PubMed
-
- Tachibana M.; Ueda J.; Fukuda M.; Takeda N.; Ohta T.; Iwanari H.; Sakihama T.; Kodama T.; Hamakubo T.; Shinkai Y. Histone Methyltransferases G9a and GLP Form Heteromeric Complexes and Are Both Crucial for Methylation of Euchromatin at H3-K9. Genes Dev. 2005, 19, 815–826. 10.1101/gad.1284005. - DOI - PMC - PubMed
-
- Peters A. H. F. M.; Kubicek S.; Mechtler K.; O’Sullivan R. J.; Derijck A. A. H. A.; Perez-Burgos L.; Kohlmaier A.; Opravil S.; Tachibana M.; Shinkai Y.; et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Mol. Cell 2003, 12, 1577–1589. 10.1016/s1097-2765(03)00477-5. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

