Optical Detection of CoV-SARS-2 Viral Proteins to Sub-Picomolar Concentrations
- PMID: 33718731
- PMCID: PMC7927290
- DOI: 10.1021/acsomega.1c00008
Optical Detection of CoV-SARS-2 Viral Proteins to Sub-Picomolar Concentrations
Abstract
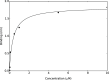
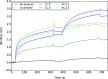
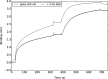
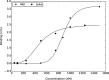
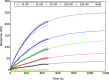
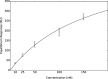
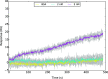
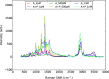
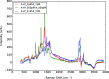
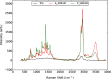
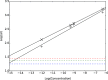
The emergence of a new strain of coronavirus in late 2019, SARS-CoV-2, led to a global pandemic in 2020. This may have been preventable if large scale, rapid diagnosis of active cases had been possible, and this has highlighted the need for more effective and efficient ways of detecting and managing viral infections. In this work, we investigate three different optical techniques for quantifying the binding of recombinant SARS-CoV-2 spike protein to surface-immobilized oligonucleotide aptamers. Biolayer interferometry is a relatively cheap, robust, and rapid method that only requires very small sample volumes. However, its detection limit of 250 nM means that it is not sensitive enough to detect antigen proteins at physiologically relevant levels (sub-pM). Surface plasmon resonance is a more sensitive technique but requires larger sample volumes, takes longer, requires more expensive instrumentation, and only reduces the detection limit to 5 nM. Surface-enhanced Raman spectroscopy is far more sensitive, enabling detection of spike protein to sub-picomolar concentrations. Control experiments performed using scrambled aptamers and using bovine serum albumin as an analyte show that this apta-sensing approach is both sensitive and selective, with no appreciable response observed for any controls. Overall, these proof-of-principle results demonstrate that SERS-based aptasensors hold great promise for development into rapid, point-of-use antigen detection systems, enabling mass testing without any need for reagents or laboratory expertise and equipment.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Boulos M. N. K.; Geraghty E. M. Geographical Tracking and Mapping of Coronavirus Disease COVID-19/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Epidemic and Associated Events Around the World: How 21st Century GIS Technologies are Supporting the Global Fight Against Outbreaks and Epidemics. Int. J. Health Geograph. 2020, 19, 8. - PMC - PubMed
-
- Jackson L. A.; Anderson E. J.; Rouphael N. G.; Roberts P. C.; Makhene M.; Coler R. N.; McCullough M. P.; Chappell J. D.; Denison M. R.; Stevens L. J.; Pruijssers A. J.; McDermott A.; Flach B.; Doria-Rose N. A.; Corbett K. S.; Morabito K. M.; O’Dell S.; Schmidt S. D.; Swanson P. A. II; Padilla M.; Mascola J. R.; Neuzil K. M.; Bennett H.; Sunn W.; Peters E.; Makowski M.; Albert J.; Cross K.; Buchanan W.; Pikaart-Tautges R.; Ledgerwood J. E.; Graham B. S.; Beigel J. H. An mRNA Vaccine Against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. 10.1056/NEJMoa2022483. - DOI - PMC - PubMed
-
- Folegatti P. M.; Ewer K. J.; Aley P. K.; Angus B.; Becker S.; Belij-Rammerstorfer S.; Bellamy D.; Bibi S.; Bittaye M.; Clutterbuck E. A.; Dold C.; Faust S. N.; Finn A.; Flaxmanm A. L.; Hallis B.; Health P.; Jenkin D.; Lazarus R.; Makinson R.; Minassian A. M.; Pollack K. M.; Ramasamy M.; Robinsin H.; Snape M.; Tarrant R.; Voysey M.; Green C.; Douglas A. D.; Hill A. V. S.; Lambe T.; Gilbert S. C.; Pollard A. J.; Safety and Immunogenicity of the ChAdOx1 nCoV-19 Vaccine Against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-blind, Randomised Controlled Trial. Lancet 2020, 396, 467–478. 10.1016/S0140-6736(20)31604-4. - DOI - PMC - PubMed
-
- Panovska-Griffiths J.; Kerr C. C.; Stuart R. M.; Mistry D.; Klein D. J.; Viner R. M.; Bonell C. Determining the Optimal Strategy for Reopening Schools, the Impact of Test and Trace Interventions, and the Risk of Occurrence of a Second COVID-19 Epidemic Wave in the UK: A Modelling Study. Lancet Child Adolesc. Health 2020, 4, 817–827. 10.1016/S2352-4642(20)30250-9. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

