Disruption of CTNND2, encoding delta-catenin, causes a penetrant attention deficit disorder and myopia
- PMID: 33718894
- PMCID: PMC7948131
- DOI: 10.1016/j.xhgg.2020.100007
Disruption of CTNND2, encoding delta-catenin, causes a penetrant attention deficit disorder and myopia
Abstract
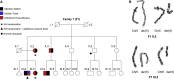
Attention deficit hyperactivity disorder (ADHD) is a common and highly heritable neurodevelopmental disorder with poorly understood pathophysiology and genetic mechanisms. A balanced chromosomal translocation interrupts CTNND2 in several members of a family with profound attentional deficit and myopia, and disruption of the gene was found in a separate unrelated individual with ADHD and myopia. CTNND2 encodes a brain-specific member of the adherens junction complex essential for postsynaptic and dendritic development, a site of potential pathophysiology in attentional disorders. Therefore, we propose that the severe and highly penetrant nature of the ADHD phenotype in affected individuals identifies CTNND2 as a potential gateway to ADHD pathophysiology similar to the DISC1 translocation in psychosis or AUTS2 in autism.
Conflict of interest statement
Declaration of interests S.P.S. and E.N. are employees and shareholders of Fulgent Genetics. J.P. is a founder of Global Gene Corp and a member of its scientific advisory board.
Figures



References
-
- Faraone S.V., Perlis R.H., Doyle A.E., Smoller J.W., Goralnick J.J., Holmgren M.A., Sklar P. Molecular genetics of attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2005;57:1313–1323. - PubMed
-
- Elia J., Gai X., Xie H.M., Perin J.C., Geiger E., Glessner J.T., D’arcy M., deBerardinis R., Frackelton E., Kim C., et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. - PMC - PubMed
-
- Stevens S.E., Sonuga-Barke E.J., Kreppner J.M., Beckett C., Castle J., Colvert E., Groothues C., Hawkins A., Rutter M. Inattention/overactivity following early severe institutional deprivation: presentation and associations in early adolescence. J. Abnorm. Child Psychol. 2008;36:385–398. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

