Two novel qualitative transcriptional signatures robustly applicable to non-research-oriented colorectal cancer samples with low-quality RNA
- PMID: 33719152
- PMCID: PMC8034468
- DOI: 10.1111/jcmm.16467
Two novel qualitative transcriptional signatures robustly applicable to non-research-oriented colorectal cancer samples with low-quality RNA
Abstract
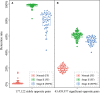
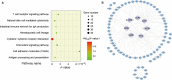
Currently, due to the low quality of RNA caused by degradation or low abundance, the accuracy of gene expression measurements by transcriptome sequencing (RNA-seq) is very challenging for non-research-oriented clinical samples, majority of which are preserved in hospitals or tissue banks worldwide with complete pathological information and follow-up data. Molecular signatures consisting of several genes are rarely applied to such samples. To utilize these resources effectively, 45 stage II non-research-oriented samples which were formalin-fixed paraffin-embedded (FFPE) colorectal carcinoma samples (CRC) using RNA-seq have been analysed. Our results showed that although gene expression measurements were significantly affected, most cancer features, based on the relative expression orderings (REOs) of gene pairs, were well preserved. We then developed two REO-based signatures, which consisted of 136 gene pairs for early diagnosis of CRC, and 4500 gene pairs for predicting post-surgery relapse risk of stage II and III CRC. The performance of our signatures, which included hundreds or thousands of gene pairs, was more robust for non-research-oriented clinical samples, compared to that of two published concise REO-based signatures. In conclusion, REO-based signatures with relatively more gene pairs could be robustly applied to non-research-oriented CRC samples.
Keywords: colorectal cancer; low-quality RNA; non-research-oriented clinical samples; relative expression orderings; transcriptional signature.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures



References
-
- Blow N. Tissue preparation: tissue issues. Nature. 2007;448(7156):959‐963. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2018Y9065/Joint Scientific and Technology Innovation Fund of Fujian Province
- 2019-WJ-32/Joint research program of health and education in Fujian Province
- 61801118/National Natural Science Foundation of China
- 81602738/National Natural Science Foundation of China
- 2020J01600/Natural Science Foundation of Fujian Province
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

