Single-Molecule FRET-Based Dynamic DNA Sensor
- PMID: 33720708
- PMCID: PMC8311664
- DOI: 10.1021/acssensors.1c00002
Single-Molecule FRET-Based Dynamic DNA Sensor
Abstract
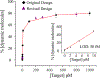
Selective and sensitive detection of nucleic acid biomarkers is of great significance in early-stage diagnosis and targeted therapy. Therefore, the development of diagnostic methods capable of detecting diseases at the molecular level in biological fluids is vital to the emerging revolution in the early diagnosis of diseases. However, the vast majority of the currently available ultrasensitive detection strategies involve either target/signal amplification or involve complex designs. Here, using a p53 tumor suppressor gene whose mutation has been implicated in more than 50% of human cancers, we show a background-free ultrasensitive detection of this gene on a simple platform. The sensor exhibits a relatively static mid-FRET state in the absence of a target that can be attributed to the time-averaged fluorescence intensity of fast transitions among multiple states, but it undergoes continuous dynamic switching between a low- and a high-FRET state in the presence of a target, allowing a high-confidence detection. In addition to its simple design, the sensor has a detection limit down to low femtomolar (fM) concentration without the need for target amplification. We also show that this sensor is highly effective in discriminating against single-nucleotide polymorphisms (SNPs). Given the generic hybridization-based detection platform, the sensing strategy developed here can be used to detect a wide range of nucleic acid sequences enabling early diagnosis of diseases and screening genetic disorders.
Keywords: DNA/RNA detection; biomarkers; fluorescence resonance energy transfer (FRET); hybridization sensor; single molecule; single-nucleotide polymorphism (SNP); ultrasensitive sensor.
Conflict of interest statement
The authors declare the following competing financial interest(s): A patent application related to this work has been filed.
Figures
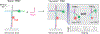


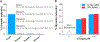
References
-
- Zhang M; Ye J; He JS; Zhang F; Ping J; Qian C; Wu J Visual detection for nucleic acid-based techniques as potential on-site detection methods. A review. Anal. Chim. Acta 2020, 1099, 1–15. - PubMed
-
- Zhao Y; Chen F; Li Q; Wang L; Fan C Isothermal Amplification of Nucleic Acids. Chem. Rev 2015, 115, 12491–12545. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

