Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome
- PMID: 33721024
- PMCID: PMC8053107
- DOI: 10.1093/nar/gkab127
Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome
Abstract
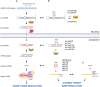
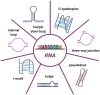
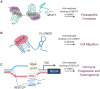
Noncoding RNAs are functional transcripts that are not translated into proteins. They represent the largest portion of the human transcriptome and have been shown to regulate gene expression networks in both physiological and pathological cell conditions. Research in this field has made remarkable progress in the comprehension of how aberrations in noncoding RNA drive relevant disease-associated phenotypes; however, the biological role and mechanism of action of several noncoding RNAs still need full understanding. Besides fulfilling its function through sequence-based mechanisms, RNA can form complex secondary and tertiary structures which allow non-canonical interactions with proteins and/or other nucleic acids. In this context, the presence of G-quadruplexes in microRNAs and long noncoding RNAs is increasingly being reported. This evidence suggests a role for RNA G-quadruplexes in controlling microRNA biogenesis and mediating noncoding RNA interaction with biological partners, thus ultimately regulating gene expression. Here, we review the state of the art of G-quadruplexes in the noncoding transcriptome, with their structural and functional characterization. In light of the existence and further possible development of G-quadruplex binders that modulate G-quadruplex conformation and protein interactions, we also discuss the therapeutic potential of G-quadruplexes as targets to interfere with disease-associated noncoding RNAs.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Hombach S., Kretz M.. Non-coding RNAs: classification, biology and functioning. Adv. Exp. Med. Biol. 2016; 937:3–17. - PubMed
-
- Cech T.R., Steitz J.A.. The noncoding RNA revolution - trashing old rules to forge new ones. Cell. 2014; 157:77–94. - PubMed
-
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011; 12:861–874. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

