SARS-CoV-2 vaccine ChAdOx1 nCoV-19 infection of human cell lines reveals low levels of viral backbone gene transcription alongside very high levels of SARS-CoV-2 S glycoprotein gene transcription
- PMID: 33722288
- PMCID: PMC7958140
- DOI: 10.1186/s13073-021-00859-1
SARS-CoV-2 vaccine ChAdOx1 nCoV-19 infection of human cell lines reveals low levels of viral backbone gene transcription alongside very high levels of SARS-CoV-2 S glycoprotein gene transcription
Abstract
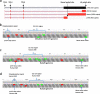
Background: ChAdOx1 nCoV-19 is a recombinant adenovirus vaccine against SARS-CoV-2 that has passed phase III clinical trials and is now in use across the globe. Although replication-defective in normal cells, 28 kbp of adenovirus genes is delivered to the cell nucleus alongside the SARS-CoV-2 S glycoprotein gene.
Methods: We used direct RNA sequencing to analyse transcript expression from the ChAdOx1 nCoV-19 genome in human MRC-5 and A549 cell lines that are non-permissive for vector replication alongside the replication permissive cell line, HEK293. In addition, we used quantitative proteomics to study over time the proteome and phosphoproteome of A549 and MRC5 cells infected with the ChAdOx1 nCoV-19 vaccine.
Results: The expected SARS-CoV-2 S coding transcript dominated in all cell lines. We also detected rare S transcripts with aberrant splice patterns or polyadenylation site usage. Adenovirus vector transcripts were almost absent in MRC-5 cells, but in A549 cells, there was a broader repertoire of adenoviral gene expression at very low levels. Proteomically, in addition to S glycoprotein, we detected multiple adenovirus proteins in A549 cells compared to just one in MRC5 cells.
Conclusions: Overall, the ChAdOx1 nCoV-19 vaccine's transcriptomic and proteomic repertoire in cell culture is as expected. The combined transcriptomic and proteomics approaches provide a detailed insight into the behaviour of this important class of vaccine using state-of-the-art techniques and illustrate the potential of this technique to inform future viral vaccine vector design.
Conflict of interest statement
SG is the co-founder of Vaccitech (co-inventors of this vaccine) and named as an inventor on a patent covering the use of ChAdOx1-vectored vaccines and a patent application covering this SARS-CoV-2 vaccine. The remaining authors declare that they have no competing interests.
Figures


References
-
- Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, Bellamy D, Bibi S, Bittaye M, Clutterbuck EA, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. - DOI - PMC - PubMed
-
- Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatullin AI, Shcheblyakov DV, Dzharullaeva AS, Grousova DM, Erokhova AS, Kovyrshina AV, Botikov AG, et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396(10255):887–97. - PMC - PubMed
-
- Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, Li JX, Yang BF, Wang L, Wang WJ, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HHSF223201510104C/U.S. Food and Drug Administration
- HHSF223201510104C/U.S. Food and Drug Administration
- BB/M02542X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M02542X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- EP/R013756/1/Engineering and Physical Sciences Research Council
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

