Igf signaling couples retina growth with body growth by modulating progenitor cell division
- PMID: 33722901
- PMCID: PMC8077508
- DOI: 10.1242/dev.199133
Igf signaling couples retina growth with body growth by modulating progenitor cell division
Abstract
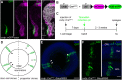
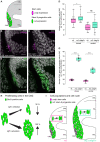
How the body and organs balance their relative growth is of key importance for coordinating size and function. This is of particular relevance in organisms, which continue to grow over their entire life span. We addressed this issue in the neuroretina of medaka fish (Oryzias latipes), a well-studied system with which to address vertebrate organ growth. We reveal that a central growth regulator, Igf1 receptor (Igf1r), is necessary and sufficient for proliferation control in the postembryonic retinal stem cell niche: the ciliary marginal zone (CMZ). Targeted activation of Igf1r signaling in the CMZ uncouples neuroretina growth from body size control, and we demonstrate that Igf1r operates on progenitor cells, stimulating their proliferation. Activation of Igf1r signaling increases retinal size while preserving its structural integrity, revealing a modular organization in which progenitor differentiation and neurogenesis are self-organized and highly regulated. Our findings position Igf signaling as a key module for controlling retinal size and composition, with important evolutionary implications.
Keywords: Medaka; Organ size; Scaling; Self-organization.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
References
-
- Attias-Geva, Z., Bentov, I., Fishman, A., Werner, H. and Bruchim, I. (2011). Insulin-like growth factor-I receptor inhibition by specific tyrosine kinase inhibitor NVP-AEW541 in endometrioid and serous papillary endometrial cancer cell lines. Gynecol. Oncol. 121, 383-389. 10.1016/j.ygyno.2011.01.008 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

