Subgenomic RNA identification in SARS-CoV-2 genomic sequencing data
- PMID: 33722935
- PMCID: PMC8015849
- DOI: 10.1101/gr.268110.120
Subgenomic RNA identification in SARS-CoV-2 genomic sequencing data
Abstract
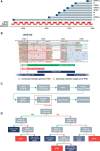
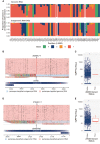
We have developed periscope, a tool for the detection and quantification of subgenomic RNA (sgRNA) in SARS-CoV-2 genomic sequence data. The translation of the SARS-CoV-2 RNA genome for most open reading frames (ORFs) occurs via RNA intermediates termed "subgenomic RNAs." sgRNAs are produced through discontinuous transcription, which relies on homology between transcription regulatory sequences (TRS-B) upstream of the ORF start codons and that of the TRS-L, which is located in the 5' UTR. TRS-L is immediately preceded by a leader sequence. This leader sequence is therefore found at the 5' end of all sgRNA. We applied periscope to 1155 SARS-CoV-2 genomes from Sheffield, United Kingdom, and validated our findings using orthogonal data sets and in vitro cell systems. By using a simple local alignment to detect reads that contain the leader sequence, we were able to identify and quantify reads arising from canonical and noncanonical sgRNA. We were able to detect all canonical sgRNAs at the expected abundances, with the exception of ORF10. A number of recurrent noncanonical sgRNAs are detected. We show that the results are reproducible using technical replicates and determine the optimum number of reads for sgRNA analysis. In VeroE6 ACE2+/- cell lines, periscope can detect the changes in the kinetics of sgRNA in orthogonal sequencing data sets. Finally, variants found in genomic RNA are transmitted to sgRNAs with high fidelity in most cases. This tool can be applied to all sequenced COVID-19 samples worldwide to provide comprehensive analysis of SARS-CoV-2 sgRNA.
© 2021 Parker et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- The Artic Network. 2020. Artic Network. https://artic.network/ncov-2019 [accessed November 5, 2020].
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
