The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis
- PMID: 33723072
- PMCID: PMC8000504
- DOI: 10.1073/pnas.2023613118
The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis
Abstract
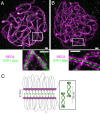
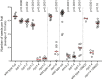
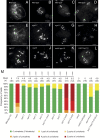
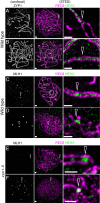
Meiotic crossovers (COs) have intriguing patterning properties, including CO interference, the tendency of COs to be well-spaced along chromosomes, and heterochiasmy, the marked difference in male and female CO rates. During meiosis, transverse filaments transiently associate the axes of homologous chromosomes, a process called synapsis that is essential for CO formation in many eukaryotes. Here, we describe the spatial organization of the transverse filaments in Arabidopsis (ZYP1) and show it to be evolutionary conserved. We show that in the absence of ZYP1 (zyp1azyp1b null mutants), chromosomes associate in pairs but do not synapse. Unexpectedly, in absence of ZYP1, CO formation is not prevented but increased. Furthermore, genome-wide analysis of recombination revealed that CO interference is abolished, with the frequent observation of close COs. In addition, heterochiasmy was erased, with identical CO rates in males and females. This shows that the tripartite synaptonemal complex is dispensable for CO formation and has a key role in regulating their number and distribution, imposing CO interference and heterochiasmy.
Keywords: crossover; heterochiasmy; interference; meiosis; synaptonemal complex.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Crossover interference: Just ZYP it.Proc Natl Acad Sci U S A. 2021 Apr 27;118(17):e2103433118. doi: 10.1073/pnas.2103433118. Proc Natl Acad Sci U S A. 2021. PMID: 33785514 Free PMC article. No abstract available.
References
-
- Muller H. J., The mechanism of crossing over II. Am. Nat. 50, 284–305 (1916).
-
- Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M., The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

