Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins
- PMID: 33723219
- PMCID: PMC7958565
- DOI: 10.1038/s41392-021-00515-5
Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins
Abstract
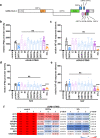
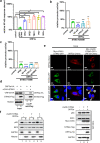
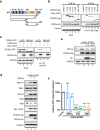
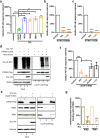
The emergence of SARS-CoV-2 has resulted in the COVID-19 pandemic, leading to millions of infections and hundreds of thousands of human deaths. The efficient replication and population spread of SARS-CoV-2 indicates an effective evasion of human innate immune responses, although the viral proteins responsible for this immune evasion are not clear. In this study, we identified SARS-CoV-2 structural proteins, accessory proteins, and the main viral protease as potent inhibitors of host innate immune responses of distinct pathways. In particular, the main viral protease was a potent inhibitor of both the RLR and cGAS-STING pathways. Viral accessory protein ORF3a had the unique ability to inhibit STING, but not the RLR response. On the other hand, structural protein N was a unique RLR inhibitor. ORF3a bound STING in a unique fashion and blocked the nuclear accumulation of p65 to inhibit nuclear factor-κB signaling. 3CL of SARS-CoV-2 inhibited K63-ubiquitin modification of STING to disrupt the assembly of the STING functional complex and downstream signaling. Diverse vertebrate STINGs, including those from humans, mice, and chickens, could be inhibited by ORF3a and 3CL of SARS-CoV-2. The existence of more effective innate immune suppressors in pathogenic coronaviruses may allow them to replicate more efficiently in vivo. Since evasion of host innate immune responses is essential for the survival of all viruses, our study provides insights into the design of therapeutic agents against SARS-CoV-2.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

