The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity
- PMID: 33723429
- PMCID: PMC7610817
- DOI: 10.1038/s41477-021-00874-5
The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity
Abstract
Plants tailor their metabolism to environmental conditions, in part through the recognition of a wide array of self and non-self molecules. In particular, the perception of microbial or plant-derived molecular patterns by cell-surface-localized pattern recognition receptors (PRRs) induces pattern-triggered immunity, which includes massive transcriptional reprogramming1. An increasing number of plant PRRs and corresponding ligands are known, but whether plants tune their immune outputs to patterns of different biological origins or of different biochemical natures remains mostly unclear. Here, we performed a detailed transcriptomic analysis in an early time series focused to study rapid-signalling transcriptional outputs induced by well-characterized patterns in the model plant Arabidopsis thaliana. This revealed that the transcriptional responses to diverse patterns (independent of their origin, biochemical nature or type of PRR) are remarkably congruent. Moreover, many of the genes most rapidly and commonly upregulated by patterns are also induced by abiotic stresses, suggesting that the early transcriptional response to patterns is part of the plant general stress response (GSR). As such, plant cells' response is in the first instance mostly to danger. Notably, the genetic impairment of the GSR reduces pattern-induced antibacterial immunity, confirming the biological relevance of this initial danger response. Importantly, the definition of a small subset of 'core immunity response' genes common and specific to pattern response revealed the function of previously uncharacterized GLUTAMATE RECEPTOR-LIKE (GLR) calcium-permeable channels in immunity. This study thus illustrates general and unique properties of early immune transcriptional reprogramming and uncovers important components of plant immunity.
Conflict of interest statement
The authors declare no competing interests.
Figures
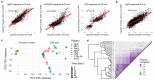
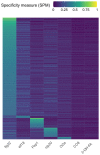
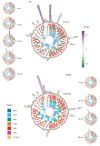

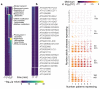
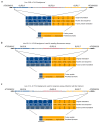
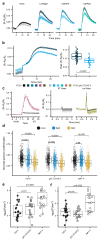
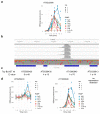


Comment in
-
Different threats, same response.Nat Plants. 2021 May;7(5):544-545. doi: 10.1038/s41477-021-00915-z. Nat Plants. 2021. PMID: 34007034 No abstract available.
References
-
- Saijo Y, Loo EP-I, Yasuda S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018;93:592–613. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

