Transcriptome array screening and verification of oral leukoplakia carcinogenesis-related hypoxia-responsive gene and microRNA
- PMID: 33723932
- PMCID: PMC7905416
- DOI: 10.7518/hxkq.2021.01.003
Transcriptome array screening and verification of oral leukoplakia carcinogenesis-related hypoxia-responsive gene and microRNA
Abstract
Objectives: To study the hypoxia response gene and microRNA (miRNA) expression profiles in the pathogenesis and progression of oral leukoplakia (OLK).
Methods: Affymetrix GeneChip human transcriptome array 2.0 was used to detect the transcriptome of normal mucosa, low-risk OLK, high-risk OLK, and early squamous cell carcinoma (SCC). Gene ontology function analysis was used to screen genes and key miRNAs whose biological role is hypoxia response. Quantitative reverse transcription polymerase ch-ain reaction (qRT-PCR) was used to verify the expression of hypoxia response genes and miRNAs.
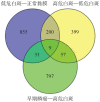
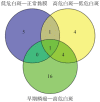
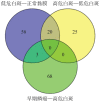
Results: A total of 7 different genes of hypoxia response between normal mucosa and low-risk OLK, 10 genes between low-risk and high-risk OLK, and 21 genes between high-risk OLK and SCC were identified. The results of qRT-PCR showed that the expression of hypoxia-inducible factor 1α, chemokine cc-motif ligand 2, and matrix metalloproteinase 3 mRNA and miR-21 in normal mucosa, OLK, and SCC increased in a stepwise manner. The expression difference between OLK and SCC was statistically significant and consistent with the results of transcriptome array.
Conclusions: The hypoxia response gene and related miRNA play roles in the development and progression of OLK.
目的: 探讨口腔白斑病癌变进程中的缺氧应答基因及相关微小RNA (miRNA)的表达。方法: 利用Affymetrix GeneChip人转录组芯片(HTA)2.0对正常口腔黏膜、低危白斑、高危白斑、口腔早期鳞癌进行转录组学检测,基因本体论(GO)功能分析筛选出生物学作用为缺氧应答的基因和相关miRNA,并采用实时定量逆转录聚合酶链反应(qRT-PCR)检测验证白斑缺氧应答基因和miRNA的表达。结果: 正常黏膜与低危白斑间有7个缺氧应答差异基因,低危白斑与高危白斑间有10个缺氧应答差异基因,高危白斑与早期鳞癌间有21个缺氧应答差异基因。缺氧应答关键基因低氧诱导因子1α、趋化因子配体2、基质金属蛋白酶3 的mRNA和miR-21在正常黏膜、口腔白斑和早期鳞癌中的表达量均呈阶梯式升高,白斑与早期鳞癌之间的表达差异均具有统计学意义,这与转录组芯片结果基本一致。结论: 缺氧应答基因及相关miRNA参与白斑病癌变。.
Keywords: hypoxia-responsive gene; microRNA; oral leukoplakia; squamous cell carcinoma.
Conflict of interest statement
利益冲突声明:作者声明本文无利益冲突。
Figures
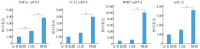
References
-
- Wang TJ, Wang L, Yang HF, et al. Development and validation of nomogram for prediction of malignant transformation in oral leukoplakia: a large-scale cohort study[J] J Oral Pathol Med. 2019;48(6):491–498. - PubMed
-
- 叶 盛. 细胞低氧感知与响应——2019年诺贝尔生理学或医学奖成果简析[J] 科技导报. 2019;37(24):41–50.
- Ye S. Cellular sensing of and responding to hypoxia: a discussion on the Nobel Prize in Physiology or Medicine 2019[J] Sci Tech Rev. 2019;37(24):41–50.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
