INO80 participates in the pathogenesis of recurrent miscarriage by epigenetically regulating trophoblast migration and invasion
- PMID: 33724648
- PMCID: PMC8051727
- DOI: 10.1111/jcmm.16322
INO80 participates in the pathogenesis of recurrent miscarriage by epigenetically regulating trophoblast migration and invasion
Abstract
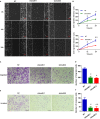
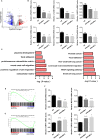
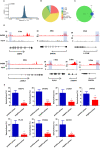
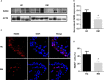
The INO80 complex, a SWI/SNF family chromatin remodeler, has regulatory effects on ESC self-renewal, somatic cell reprogramming and blastocyst development. However, the role of INO80 in regulating trophoblast cells and recurrent miscarriage (RM) remains elusive. To investigate the in vivo effects of Ino80 in embryo development, we disrupted Ino80 in C57 mice, which resulted in embryonic lethality. Silencing of Ino80 led to decreased survival capacity, migration and invasion of trophoblasts. Furthermore, RNA high-throughput sequencing (RNA-seq) revealed that Ino80 silencing closely resembled the gene expression changes in RM tissues. To investigate the mechanisms for these results, RNA-seq combined with high-throughput sequencing (ChIP-seq) was used in trophoblast cells, and it showed that Ino80 physically occupies promoter regions to affect the expression of invasion-associated genes. Last, Western blotting analyses and immunofluorescence staining revealed that the content of INO80 was reduced in RM patients compared to in healthy controls. This study indicates that INO80 has a specific regulatory effect on the viability, migration and invasion of trophoblast cells. Combined with its regulation of the expression of invasion-associated genes, it has been proposed that epigenetic regulation plays an important role in the occurrence of RM, potentially informing RM therapeutic strategies.
Keywords: INO80; invasive; recurrent miscarriage; trophoblast.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures


Similar articles
-
The miR-27a-3p/USP25 axis participates in the pathogenesis of recurrent miscarriage by inhibiting trophoblast migration and invasion.J Cell Physiol. 2019 Nov;234(11):19951-19963. doi: 10.1002/jcp.28593. Epub 2019 Apr 5. J Cell Physiol. 2019. PMID: 30953360
-
MNSFβ Promotes the Proliferation and Migration of Human Extravillous Trophoblast Cells and the Villus Expression Level of MNSFβ Is Decreased in Recurrent Miscarriage Patients.Gynecol Obstet Invest. 2021;86(1-2):27-39. doi: 10.1159/000506309. Epub 2020 Dec 16. Gynecol Obstet Invest. 2021. PMID: 33326956
-
KLF4 regulates trophoblast function and associates with unexplained recurrent spontaneous abortion.J Transl Med. 2024 Oct 10;22(1):922. doi: 10.1186/s12967-024-05707-5. J Transl Med. 2024. PMID: 39390495 Free PMC article.
-
Traditional and New Routes of Trophoblast Invasion and Their Implications for Pregnancy Diseases.Int J Mol Sci. 2019 Dec 31;21(1):289. doi: 10.3390/ijms21010289. Int J Mol Sci. 2019. PMID: 31906245 Free PMC article. Review.
-
The motile and invasive capacity of human endometrial stromal cells: implications for normal and impaired reproductive function.Hum Reprod Update. 2013 Sep-Oct;19(5):542-57. doi: 10.1093/humupd/dmt025. Epub 2013 Jul 4. Hum Reprod Update. 2013. PMID: 23827985 Review.
Cited by
-
INO80 Is Required for the Cell Cycle Control, Survival, and Differentiation of Mouse ESCs by Transcriptional Regulation.Int J Mol Sci. 2022 Dec 6;23(23):15402. doi: 10.3390/ijms232315402. Int J Mol Sci. 2022. PMID: 36499727 Free PMC article.
-
Epigenetic drivers of metalloproteinases and metastasis.Trends Cell Biol. 2025 Mar 14:S0962-8924(25)00044-3. doi: 10.1016/j.tcb.2025.02.010. Online ahead of print. Trends Cell Biol. 2025. PMID: 40089451 Review.
References
-
- Norwitz ER, Schust DJ, Fisher SJ. Implantation and the survival of early pregnancy. N Engl J Med. 2001;345(19):1400‐1408. - PubMed
-
- Dwi Putra SE, Reichetzeder C, Meixner M, Liere K, Slowinski T, Hocher B. DNA methylation of the glucocorticoid receptor gene promoter in the placenta is associated with blood pressure regulation in human pregnancy. J Hypertens. 2017;35(11):2276‐2286. - PubMed
-
- Practice Committee of the American Society for Reproductive M . Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril. 2012;98(5):1103‐1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

