Transcriptional profiling of the response to the trichloroethylene metabolite S-(1,2-dichlorovinyl)-L-cysteine revealed activation of the eIF2α/ATF4 integrated stress response in two in vitro placental models
- PMID: 33725128
- PMCID: PMC7961173
- DOI: 10.1007/s00204-021-03011-5
Transcriptional profiling of the response to the trichloroethylene metabolite S-(1,2-dichlorovinyl)-L-cysteine revealed activation of the eIF2α/ATF4 integrated stress response in two in vitro placental models
Abstract
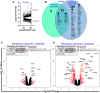
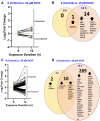
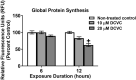
Trichloroethylene (TCE) is an industrial solvent and widespread environmental contaminant. Although TCE exposure is prevalent, epidemiological studies of TCE exposure associations with adverse birth outcomes are inconclusive. Prior studies show that the TCE metabolite S-(1,2-dichlorovinyl)-L-cysteine (DCVC) exhibits toxicity in a placental cell line. In the current study, genome-wide gene expression and gene set enrichment analyses were used to identify novel genes and pathway alterations in the HTR-8/SVneo human trophoblast cell line and human placental villous explants treated with DCVC at concentrations relevant to human exposures. In the cells, concentration- and time-dependent effects were observed, as evidenced by the magnitude of altered gene expression after treatment with 20 µM DCVC versus 10 µM, and 12-h versus 6-h of treatment. Comparing the two models for the transcriptional response to 12-h 20 µM DCVC treatment, no differentially expressed genes reached significance in villous explants, whereas 301 differentially expressed genes were detected in HTR-8/SVneo cells compared with non-treated controls (FDR < 0.05 + LogFC > 0.35 [FC > 1.3]). GSEA revealed five upregulated enriched pathways in common between explants and cells (FDR < 0.05). Moreover, all 12-h DCVC treatment groups from both models contained upregulated pathways enriched for genes regulated by the ATF4 transcription factor. The overrepresentation of ATF4 regulation of differentially expressed genes indicated activation of the integrated stress response (ISR), a condition triggered by multiple stress stimuli, including the unfolded protein response. DCVC-induced ISR activation was confirmed by elevated eIF2α phosphorylation, ATF4 protein concentrations, and decreased global protein synthesis in HTR-8/SVneo cells. This study identifies a mechanism of DCVC-induced cytotoxicity by revealing the involvement of a specific stress signaling pathway.
Keywords: ATF4; Integrated stress response; Placenta; S-(1,2-dichlorovinyl)-l-cysteine (DCVC); Trichloroethylene.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures











Similar articles
-
Reactive Oxygen Stimulation of Interleukin-6 Release in the Human Trophoblast Cell Line HTR-8/SVneo by the Trichlorethylene Metabolite S-(1,2-Dichloro)-l-Cysteine.Biol Reprod. 2016 Sep;95(3):66. doi: 10.1095/biolreprod.116.139261. Epub 2016 Aug 3. Biol Reprod. 2016. PMID: 27488030 Free PMC article.
-
The trichloroethylene metabolite S-(1,2-dichlorovinyl)-L-cysteine induces progressive mitochondrial dysfunction in HTR-8/SVneo trophoblasts.Toxicology. 2019 Nov 1;427:152283. doi: 10.1016/j.tox.2019.152283. Epub 2019 Aug 30. Toxicology. 2019. PMID: 31476333 Free PMC article.
-
Exposure to Trichloroethylene Metabolite S-(1,2-Dichlorovinyl)-L-cysteine Causes Compensatory Changes to Macronutrient Utilization and Energy Metabolism in Placental HTR-8/SVneo Cells.Chem Res Toxicol. 2020 Jun 15;33(6):1339-1355. doi: 10.1021/acs.chemrestox.9b00356. Epub 2020 Jan 30. Chem Res Toxicol. 2020. PMID: 31951115 Free PMC article.
-
Renal toxicity and carcinogenicity of trichloroethylene: key results, mechanisms, and controversies.Crit Rev Toxicol. 2000 May;30(3):253-85. doi: 10.1080/10408440091159202. Crit Rev Toxicol. 2000. PMID: 10852497 Review.
-
Mutagenicity of trichloroethylene and its metabolites: implications for the risk assessment of trichloroethylene.Environ Health Perspect. 2000 May;108 Suppl 2(Suppl 2):215-23. doi: 10.1289/ehp.00108s2215. Environ Health Perspect. 2000. PMID: 10807553 Free PMC article. Review.
Cited by
-
Transcriptomic comparison of in vitro models of the human placenta.bioRxiv [Preprint]. 2024 Jun 16:2024.06.14.598695. doi: 10.1101/2024.06.14.598695. bioRxiv. 2024. Update in: Placenta. 2025 Jan;159:52-61. doi: 10.1016/j.placenta.2024.11.007. PMID: 38915703 Free PMC article. Updated. Preprint.
-
Sexually concordant and dimorphic transcriptional responses to maternal trichloroethylene and/or N-acetyl cysteine exposure in Wistar rat placental tissue.Toxicology. 2023 Jan 1;483:153371. doi: 10.1016/j.tox.2022.153371. Epub 2022 Nov 14. Toxicology. 2023. PMID: 36396003 Free PMC article.
-
Placental single cell transcriptomics: Opportunities for endocrine disrupting chemical toxicology.Mol Cell Endocrinol. 2023 Dec 1;578:112066. doi: 10.1016/j.mce.2023.112066. Epub 2023 Sep 9. Mol Cell Endocrinol. 2023. PMID: 37690473 Free PMC article.
-
A transcriptomic comparison of in vitro models of the human placenta.Placenta. 2025 Jan;159:52-61. doi: 10.1016/j.placenta.2024.11.007. Epub 2024 Nov 26. Placenta. 2025. PMID: 39637677 Free PMC article.
-
Current approaches and advances in placental toxicology.Trends Endocrinol Metab. 2025 Jun 4:S1043-2760(25)00100-6. doi: 10.1016/j.tem.2025.05.001. Online ahead of print. Trends Endocrinol Metab. 2025. PMID: 40473500 Review.
References
-
- Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. In: Bahraham Bioinformaticz. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 27 Feb 2019
-
- ATSDR (2016) Public Health Statement on Trichloroethylene. In: Agency for Toxic Substances and Disease Registry. https://www.atsdr.cdc.gov/ToxProfiles/tp19-c1.pdf Accessed 4 Nov 2019
-
- ATCC (2015) American Type Culture Collection Product Sheet: HTR8/SVneo (ATCC® CRL3271™). American Type Culture Collection
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

