Sequence-Selective Protection of Peptides from Proteolysis
- PMID: 33725413
- PMCID: PMC8252432
- DOI: 10.1002/anie.202102148
Sequence-Selective Protection of Peptides from Proteolysis
Abstract
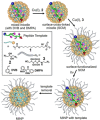
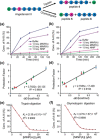
Proteolysis of proteins and peptides is involved in the infection of cells by enveloped viruses and also in the invasion and spread of cancer cells. Shutting down broad-specificity proteases, however, is problematic because normal functions by these proteases will be affected. Herein, nanoparticle receptors were prepared from molecular imprinting for complex biological peptides. Their strong and selective binding enabled them to protect their targeted sequences from proteolysis in aqueous solution at stoichiometric amounts. Generality of the method was demonstrated by the protection of hydrophobic and hydrophilic peptides from different proteases, selective protection of a segment of a long peptide, and selective protection of a targeted peptide in a mixture. Most interestingly, two receptors targeting different parts of a long peptide could work in cooperation to protect the overall sequence, highlighting the versatility of the method.
Keywords: inhibition; molecular imprinting; peptide; protease; protection.
© 2021 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Substrate Protection in Controlled Enzymatic Transformation of Peptides and Proteins.Chembiochem. 2021 Sep 2;22(17):2680-2687. doi: 10.1002/cbic.202100217. Epub 2021 Jun 14. Chembiochem. 2021. PMID: 34058051 Free PMC article.
-
Peptide Backbone Composition and Protease Susceptibility: Impact of Modification Type, Position, and Tandem Substitution.Chembiochem. 2016 Apr 15;17(8):712-8. doi: 10.1002/cbic.201500312. Epub 2015 Aug 20. Chembiochem. 2016. PMID: 26205791 Free PMC article.
-
Circulating Peptidome and Tumor-Resident Proteolysis.Enzymes. 2017;42:1-25. doi: 10.1016/bs.enz.2017.08.001. Epub 2017 Oct 10. Enzymes. 2017. PMID: 29054266 Review.
-
Prediction of Proteases Involved in Peptide Generation.Methods Mol Biol. 2017;1574:205-213. doi: 10.1007/978-1-4939-6850-3_15. Methods Mol Biol. 2017. PMID: 28315253
-
N- and C-terminal degradomics: new approaches to reveal biological roles for plant proteases from substrate identification.Physiol Plant. 2012 May;145(1):5-17. doi: 10.1111/j.1399-3054.2011.01536.x. Epub 2011 Dec 7. Physiol Plant. 2012. PMID: 22023699 Review.
Cited by
-
Molecularly imprinted materials for glycan recognition and processing.J Mater Chem B. 2022 Sep 15;10(35):6607-6617. doi: 10.1039/d2tb00164k. J Mater Chem B. 2022. PMID: 35481837 Free PMC article. Review.
-
Molecular imprinting of glycoproteins: From preparation to cancer theranostics.Theranostics. 2022 Feb 28;12(5):2406-2426. doi: 10.7150/thno.69189. eCollection 2022. Theranostics. 2022. PMID: 35265217 Free PMC article. Review.
-
Substrate Protection in Controlled Enzymatic Transformation of Peptides and Proteins.Chembiochem. 2021 Sep 2;22(17):2680-2687. doi: 10.1002/cbic.202100217. Epub 2021 Jun 14. Chembiochem. 2021. PMID: 34058051 Free PMC article.
-
Epitope-imprinted polymers: Design principles of synthetic binding partners for natural biomacromolecules.Sci Adv. 2021 Oct 29;7(44):eabi9884. doi: 10.1126/sciadv.abi9884. Epub 2021 Oct 29. Sci Adv. 2021. PMID: 34714673 Free PMC article.
-
Amyloid-reoriented enzyme catalysis.Nat Commun. 2025 Apr 2;16(1):3164. doi: 10.1038/s41467-025-58536-5. Nat Commun. 2025. PMID: 40175427 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

