Single-Cell RNA Sequencing Uncovers Paracrine Functions of the Epicardial-Derived Cells in Arrhythmogenic Cardiomyopathy
- PMID: 33726497
- PMCID: PMC8169643
- DOI: 10.1161/CIRCULATIONAHA.120.052928
Single-Cell RNA Sequencing Uncovers Paracrine Functions of the Epicardial-Derived Cells in Arrhythmogenic Cardiomyopathy
Abstract
Background: Arrhythmogenic cardiomyopathy (ACM) manifests with sudden death, arrhythmias, heart failure, apoptosis, and myocardial fibro-adipogenesis. The phenotype typically starts at the epicardium and advances transmurally. Mutations in genes encoding desmosome proteins, including DSP (desmoplakin), are major causes of ACM.
Methods: To delineate contributions of the epicardium to the pathogenesis of ACM, the Dsp allele was conditionally deleted in the epicardial cells in mice upon expression of tamoxifen-inducible Cre from the Wt1 locus. Wild type (WT) and Wt1-CreERT2:DspW/F were crossed to Rosa26mT/mG (R26mT/mG) dual reporter mice to tag the epicardial-derived cells with the EGFP (enhanced green fluorescent protein) reporter protein. Tagged epicardial-derived cells from adult Wt1-CreERT2:R26mT/mG and Wt1-CreERT2: R26mT/mG:DspW/F mouse hearts were isolated by fluorescence-activated cell staining and sequenced by single-cell RNA sequencing.
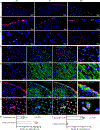
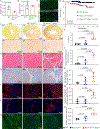
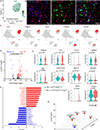
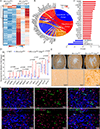
Results: WT1 (Wilms tumor 1) expression was progressively restricted postnatally and was exclusive to the epicardium by postnatal day 21. Expression of Dsp was reduced in the epicardial cells but not in cardiac myocytes in the Wt1-CreERT2:DspW/F mice. The Wt1-CreERT2:DspW/F mice exhibited premature death, cardiac dysfunction, arrhythmias, myocardial fibro-adipogenesis, and apoptosis. Single-cell RNA sequencing of ≈18 000 EGFP-tagged epicardial-derived cells identified genotype-independent clusters of endothelial cells, fibroblasts, epithelial cells, and a very small cluster of cardiac myocytes, which were confirmed on coimmunofluorescence staining of the myocardial sections. Differentially expressed genes between the paired clusters in the 2 genotypes predicted activation of the inflammatory and mitotic pathways-including the TGFβ1 (transforming growth factor β1) and fibroblast growth factors-in the epicardial-derived fibroblast and epithelial clusters, but predicted their suppression in the endothelial cell cluster. The findings were corroborated by analysis of gene expression in the pooled RNA-sequencing data, which identified predominant dysregulation of genes involved in epithelial-mesenchymal transition, and dysregulation of 146 genes encoding the secreted proteins (secretome), including genes in the TGFβ1 pathway. Activation of the TGFβ1 and its colocalization with fibrosis in the Wt1-CreERT2:R26mT/mG:DspW/F mouse heart was validated by complementary methods.
Conclusions: Epicardial-derived cardiac fibroblasts and epithelial cells express paracrine factors, including TGFβ1 and fibroblast growth factors, which mediate epithelial-mesenchymal transition, and contribute to the pathogenesis of myocardial fibrosis, apoptosis, arrhythmias, and cardiac dysfunction in a mouse model of ACM. The findings uncover contributions of the epicardial-derived cells to the pathogenesis of ACM.
Keywords: arrhythmias, cardiac; paracrine communication; pericardium; sequence analysis, RNA.
Conflict of interest statement
Disclosure
The authors declare that there is no actual or perceived potential conflict of interest and there has been none during the last two years.
Figures




Similar articles
-
Exercise restores dysregulated gene expression in a mouse model of arrhythmogenic cardiomyopathy.Cardiovasc Res. 2020 May 1;116(6):1199-1213. doi: 10.1093/cvr/cvz199. Cardiovasc Res. 2020. PMID: 31350552 Free PMC article.
-
Pharmacological suppression of the WNT signaling pathway attenuates age-dependent expression of the phenotype in a mouse model of arrhythmogenic cardiomyopathy.J Cardiovasc Aging. 2021;1(3):10.20517/jca.2021.04. doi: 10.20517/jca.2021.04. Epub 2021 Jun 6. J Cardiovasc Aging. 2021. PMID: 34447973 Free PMC article.
-
PANoptosis is a prominent feature of desmoplakin cardiomyopathy.J Cardiovasc Aging. 2023 Feb;3(1):3. doi: 10.20517/jca.2022.34. Epub 2023 Jan 1. J Cardiovasc Aging. 2023. PMID: 36818425 Free PMC article.
-
Established and Emerging Mechanisms in the Pathogenesis of Arrhythmogenic Cardiomyopathy: A Multifaceted Disease.Int J Mol Sci. 2020 Aug 31;21(17):6320. doi: 10.3390/ijms21176320. Int J Mol Sci. 2020. PMID: 32878278 Free PMC article. Review.
-
Desmoplakin Cardiomyopathy: Comprehensive Review of an Increasingly Recognized Entity.J Clin Med. 2023 Apr 3;12(7):2660. doi: 10.3390/jcm12072660. J Clin Med. 2023. PMID: 37048743 Free PMC article. Review.
Cited by
-
Exploring the Function of Epicardial Cells Beyond the Surface.Circ Res. 2024 Jul 5;135(2):353-371. doi: 10.1161/CIRCRESAHA.124.321567. Epub 2024 Jul 4. Circ Res. 2024. PMID: 38963865 Free PMC article. Review.
-
Integrative Single-Cell and Spatial Transcriptomics Reveal Functional and Spatial Heterogeneity of Atrial and Ventricular Cardiomyocytes in the Heart.Mol Biotechnol. 2025 Jun 9. doi: 10.1007/s12033-025-01443-3. Online ahead of print. Mol Biotechnol. 2025. PMID: 40488964
-
Recent progress of iPSC technology in cardiac diseases.Arch Toxicol. 2021 Dec;95(12):3633-3650. doi: 10.1007/s00204-021-03172-3. Epub 2021 Oct 17. Arch Toxicol. 2021. PMID: 34657219 Free PMC article. Review.
-
AAV-Mediated Delivery of Plakophilin-2a Arrests Progression of Arrhythmogenic Right Ventricular Cardiomyopathy in Murine Hearts: Preclinical Evidence Supporting Gene Therapy in Humans.Circ Genom Precis Med. 2024 Feb;17(1):e004305. doi: 10.1161/CIRCGEN.123.004305. Epub 2024 Jan 30. Circ Genom Precis Med. 2024. PMID: 38288614 Free PMC article.
-
Maturation of human cardiac organoids enables complex disease modeling and drug discovery.Nat Cardiovasc Res. 2025 Jul;4(7):821-840. doi: 10.1038/s44161-025-00669-3. Epub 2025 Jun 25. Nat Cardiovasc Res. 2025. PMID: 40562874 Free PMC article.
References
-
- Corrado D, Basso C and Judge DP. Arrhythmogenic Cardiomyopathy. Circ Res. 2017;121:784–802. - PubMed
-
- Gandjbakhch E, Redheuil A, Pousset F, Charron P and Frank R. Clinical Diagnosis, Imaging, and Genetics of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia: JACC State-of-the-Art Review. J Am Coll Cardiol. 2018;72:784–804. - PubMed
-
- Finocchiaro G, Papadakis M, Robertus JL, Dhutia H, Steriotis AK, Tome M, Mellor G, Merghani A, Malhotra A, Behr E, et al. Etiology of Sudden Death in Sports: Insights From a United Kingdom Regional Registry. J Am Coll Cardiol. 2016;67:2108–2115. - PubMed
-
- Thiene G, Nava A, Corrado D, Rossi L and Pennelli N. Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med. 1988;318:129–133. - PubMed
-
- Corrado D, Basso C, Thiene G, McKenna WJ, Davies MJ, Fontaliran F, Nava A, Silvestri F, Blomstrom-Lundqvist C, Wlodarska EK, et al. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: a multicenter study. J Am Coll Cardiol. 1997;30:1512–1520. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

