A next-generation sequencing-based strategy combining microsatellite instability and tumor mutation burden for comprehensive molecular diagnosis of advanced colorectal cancer
- PMID: 33726687
- PMCID: PMC7962287
- DOI: 10.1186/s12885-021-07942-1
A next-generation sequencing-based strategy combining microsatellite instability and tumor mutation burden for comprehensive molecular diagnosis of advanced colorectal cancer
Abstract
Background: Mismatch repair (MMR)/microsatellite instability (MSI) and tumor mutational burden (TMB) are independent biomarkers that complement each other for predicting immune checkpoint inhibitors (ICIs) efficacy. Here we aim to establish a strategy that integrates MSI and TMB determination for colorectal cancer (CRC) in one single assay.
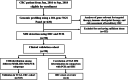
Methods: Surgical or biopsy specimens retrospectively collected from CRC patients were subjected to NGS analysis. Immunohistochemistry (IHC) and polymerase chain reaction (PCR) were also used to determine MMR/MSI for those having enough tissues. The NGS-MSI method was validated against IHC and PCR. The MSI-high (MSI-H) or microsatellite stable (MSS) groups were further stratified based on tumor mutational burden, followed by validation using the The Cancer Genome Atlas (TCGA) CRC dataset. Immune microenvironment was evaluated for each subgroup be profiling the expression of immune signatures.
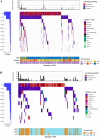
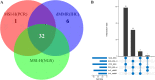
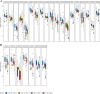
Results: Tissues from 430 CRC patients were analyzed using a 381-gene NGS panel. Alterations in KRAS, NRAS, BRAF, and HER2 occurred at a significantly higher incidence among MSI-H tumors than in MSS patients (83.6% vs. 58.4%, p = 0.0003). A subset comprising 98 tumors were tested for MSI/MMR using all three techniques, where NGS proved to be 99.0 and 93.9% concordant with PCR and IHC, respectively. Four of the 7 IHC-PCR discordant cases had low TMB (1.1-8.1 muts/Mb) and were confirmed to have been misdiagnosed by IHC. Intriguingly, 4 of the 66 MSS tumors (as determined by NGS) were defined as TMB-high (TMB-H) using a cut-off of 29 mut/Mb. Likewise, 15 of the 456 MSS tumors in the TCGA CRC cohort were also TMB-H with a cut-off of 9 muts/Mb. Expression of immune signatures across subgroups (MSS-TMB-H, MSI-H-TMB-H, and MSS-TMB-L) confirmed that the microenvironment of the MSS-TMB-H tumors was similar to that of the MSI-H-TMB-H tumors, but significantly more immune-responsive than that of the MSS-TMB-L tumors, indicating that MSI combined with TMB may be more precise than MSI alone for immune microenvironment prediction.
Conclusion: This study demonstrated that NGS panel-based method is both robust and tissue-efficient for comprehensive molecular diagnosis of CRC. It also underscores the importance of combining MSI and TMB information for discerning patients with different microenvironment.
Keywords: Colorectal cancer; Immune checkpoint inhibitor; Microsatellite instability; Next generation sequencing; Tumor mutation burden.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Gupta S, Provenzale D, Llor X, Halverson AL, Grady W, Chung DC, Haraldsdottir S, Markowitz AJ, Slavin TP, Jr, Hampel H, et al. NCCN guidelines insights: genetic/familial high-risk assessment: colorectal, version 2.2019. J Natl Compr Cancer Netw. 2019;17(9):1032–1041. doi: 10.6004/jnccn.2019.0044. - DOI - PubMed
-
- Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

