Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients
- PMID: 33726816
- PMCID: PMC7968334
- DOI: 10.1186/s13073-021-00855-5
Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients
Abstract
Background: We report the findings from 4437 individuals (3219 patients and 1218 relatives) who have been analyzed by whole genome sequencing (WGS) at the Genomic Medicine Center Karolinska-Rare Diseases (GMCK-RD) since mid-2015. GMCK-RD represents a long-term collaborative initiative between Karolinska University Hospital and Science for Life Laboratory to establish advanced, genomics-based diagnostics in the Stockholm healthcare setting.
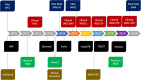
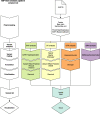
Methods: Our analysis covers detection and interpretation of SNVs, INDELs, uniparental disomy, CNVs, balanced structural variants, and short tandem repeat expansions. Visualization of results for clinical interpretation is carried out in Scout-a custom-developed decision support system. Results from both singleton (84%) and trio/family (16%) analyses are reported. Variant interpretation is done by 15 expert teams at the hospital involving staff from three clinics. For patients with complex phenotypes, data is shared between the teams.
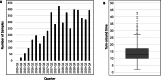
Results: Overall, 40% of the patients received a molecular diagnosis ranging from 19 to 54% for specific disease groups. There was heterogeneity regarding causative genes (n = 754) with some of the most common ones being COL2A1 (n = 12; skeletal dysplasia), SCN1A (n = 8; epilepsy), and TNFRSF13B (n = 4; inborn errors of immunity). Some causative variants were recurrent, including previously known founder mutations, some novel mutations, and recurrent de novo mutations. Overall, GMCK-RD has resulted in a large number of patients receiving specific molecular diagnoses. Furthermore, negative cases have been included in research studies that have resulted in the discovery of 17 published, novel disease-causing genes. To facilitate the discovery of new disease genes, GMCK-RD has joined international data sharing initiatives, including ClinVar, UDNI, Beacon, and MatchMaker Exchange.
Conclusions: Clinical WGS at GMCK-RD has provided molecular diagnoses to over 1200 individuals with a broad range of rare diseases. Consolidation and spread of this clinical-academic partnership will enable large-scale national collaboration.
Keywords: Clinical diagnostics; Monogenic disease; Single nucleotide variant; Whole genome sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- INSERM. Orphanet: an online database of rare diseases and orphan drugs. http://www.orpha.net. Accessed 31st Dec 2020.
-
- McKusick-Nathans Institute of Genetic Medicine JHUB, MD). Online Mendelian Inheritance in Man, OMIM®. https://omim.org/. Accessed 31st Dec 2020.
-
- EURORDIS. What is a rare disease? https://www.eurordis.org/content/what-rare-disease. Accessed 31st Dec 2020.
-
- Vrueh RD, Baekelandt ERF, Haan JMHD. Background Paper 6.19 Rare Diseases. Priority Medicines for Europe and the World “A Public Health Approach to Innovation”. 2013.
-
- Olry A, Rath A. Prevalence of rare diseases: bibliographic data. Orphanet report series, Rare diseases collection. 2015.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

