Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus
- PMID: 33727169
- PMCID: PMC7955574
- DOI: 10.1016/j.micpath.2021.104831
Mutations in SARS-CoV-2; Consequences in structure, function, and pathogenicity of the virus
Abstract
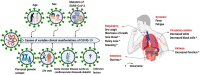
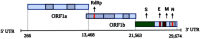
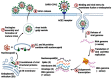
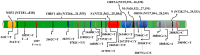
The third pandemic of coronavirus infection, called COVID-19 disease, began recently in China. The newly discovered coronavirus, entitled SARS-CoV-2, is the seventh member of the human coronaviruses. The main pathogenesis of SARS-CoV-2 infection is severe pneumonia, RNAaemia, accompanied by glass turbidity, and acute cardiac injury. It possesses a single-stranded positive-sense RNA genome which is 60-140 nm in diameter, and has a size of 26-32 kbp. Viral pathogenesis is accomplished with spike glycoprotein through the employment of a membrane-bound aminopeptidase, called the ACE2, as its primary cell receptor. It has been confirmed that various factors such as different national rules for quarantine and various races or genetic backgrounds might influence the mortality and infection rate of COVID-19 in the geographic areas. In addition to various known and unknown factors and host genetic susceptibility, mutations and genetic variabilities of the virus itself have a critical impact on variable clinical features of COVID-19. Although the SARS-CoV-2 genome is more stable than SARS-CoV or MERS-CoV, it has a relatively high dynamic mutation rate with respect to other RNA viruses. It's noteworthy that, some mutations can be founder mutations and show specific geographic patterns. Undoubtedly, these mutations can drive viral genetic variability, and because of genotype-phenotype correlation, resulting in a virus with more/lower/no decrease in natural pathogenic fitness or on the other scenario, facilitating their rapid antigenic shifting to escape the host immunity and also inventing a drug resistance virus, so converting it to a more infectious or deadly virus. Overall, the detection of all mutations in SARS-CoV-2 and their relations with pathological changes is nearly impossible, mostly due to asymptomatic subjects. In this review paper, the reported mutations of the SARS-CoV-2 and related variations in virus structure and pathogenicity in different geographic areas and genotypes are widely investigated. Many studies need to be repeated in other regions/locations for other people to confirm the findings. Such studies could benefit patient-specific therapy, according to genotyping patterns of SARS-CoV-2 distribution.
Keywords: COVID-19; Mutation; Pandemic disease; Pathogenicity; SARS-CoV-2.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

