One versus Many: Polymicrobial Communities and the Cystic Fibrosis Airway
- PMID: 33727344
- PMCID: PMC8092191
- DOI: 10.1128/mBio.00006-21
One versus Many: Polymicrobial Communities and the Cystic Fibrosis Airway
Abstract
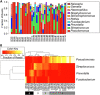
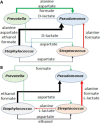
Culture-independent studies have revealed that chronic lung infections in persons with cystic fibrosis (pwCF) are rarely limited to one microbial species. Interactions among bacterial members of these polymicrobial communities in the airways of pwCF have been reported to modulate clinically relevant phenotypes. Furthermore, it is clear that a single polymicrobial community in the context of CF airway infections cannot explain the diversity of clinical outcomes. While large 16S rRNA gene-based studies have allowed us to gain insight into the microbial composition and predicted functional capacities of communities found in the CF lung, here we argue that in silico approaches can help build clinically relevant in vitro models of polymicrobial communities that can in turn be used to experimentally test and validate computationally generated hypotheses. Furthermore, we posit that combining computational and experimental approaches will enhance our understanding of mechanisms that drive microbial community function and identify new therapeutics to target polymicrobial infections.
Keywords: antibiotics; biofilms; chronic infection; cystic fibrosis; metabolic modeling; microbial communities; microbiome.
Copyright © 2021 Jean-Pierre et al.
Figures
Similar articles
-
Bacterial interactions underpin worsening lung function in cystic fibrosis-associated infections.mBio. 2025 Jan 8;16(1):e0145624. doi: 10.1128/mbio.01456-24. Epub 2024 Nov 22. mBio. 2025. PMID: 39576107 Free PMC article.
-
Disruption of Cross-Feeding Inhibits Pathogen Growth in the Sputa of Patients with Cystic Fibrosis.mSphere. 2020 Apr 29;5(2):e00343-20. doi: 10.1128/mSphere.00343-20. mSphere. 2020. PMID: 32350096 Free PMC article.
-
Dpr-mediated H2O2 resistance contributes to streptococcus survival in a cystic fibrosis airway model system.J Bacteriol. 2024 Jul 25;206(7):e0017624. doi: 10.1128/jb.00176-24. Epub 2024 Jun 28. J Bacteriol. 2024. PMID: 38940597 Free PMC article.
-
The polymicrobial nature of airway infections in cystic fibrosis: Cangene Gold Medal Lecture.Can J Microbiol. 2011 Feb;57(2):69-77. doi: 10.1139/w10-105. Can J Microbiol. 2011. PMID: 21326348 Review.
-
The Yin and Yang of Streptococcus Lung Infections in Cystic Fibrosis: a Model for Studying Polymicrobial Interactions.J Bacteriol. 2019 May 8;201(11):e00115-19. doi: 10.1128/JB.00115-19. Print 2019 Jun 1. J Bacteriol. 2019. PMID: 30885933 Free PMC article. Review.
Cited by
-
Bacterial interactions underpin worsening lung function in cystic fibrosis-associated infections.mBio. 2025 Jan 8;16(1):e0145624. doi: 10.1128/mbio.01456-24. Epub 2024 Nov 22. mBio. 2025. PMID: 39576107 Free PMC article.
-
Pseudomonas aeruginosa surface motility and invasion into competing communities enhance interspecies antagonism.mBio. 2024 Sep 11;15(9):e0095624. doi: 10.1128/mbio.00956-24. Epub 2024 Aug 6. mBio. 2024. PMID: 39105585 Free PMC article.
-
Antibacterial potential of Stenotrophomonas maltophilia complex cystic fibrosis isolates.mSphere. 2024 Jul 30;9(7):e0033524. doi: 10.1128/msphere.00335-24. Epub 2024 Jul 9. mSphere. 2024. PMID: 38980073 Free PMC article.
-
Community composition shapes microbial-specific phenotypes in a cystic fibrosis polymicrobial model system.Elife. 2023 Jan 20;12:e81604. doi: 10.7554/eLife.81604. Elife. 2023. PMID: 36661299 Free PMC article.
-
Distribution and diversity of type VI secretion system clusters in Enterobacter bugandensis and Enterobacter cloacae.Microb Genom. 2023 Dec;9(12):001148. doi: 10.1099/mgen.0.001148. Microb Genom. 2023. PMID: 38054968 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

