Critical ACE2 Determinants of SARS-CoV-2 and Group 2B Coronavirus Infection and Replication
- PMID: 33727353
- PMCID: PMC8092278
- DOI: 10.1128/mBio.03149-20
Critical ACE2 Determinants of SARS-CoV-2 and Group 2B Coronavirus Infection and Replication
Abstract
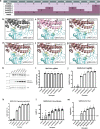
The angiotensin-converting enzyme 2 (ACE2) receptor is a major severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) host range determinant, and understanding SARS-CoV-2-ACE2 interactions will provide important insights into COVID-19 pathogenesis and animal model development. SARS-CoV-2 cannot infect mice due to incompatibility between its receptor binding domain and the murine ACE2 receptor. Through molecular modeling and empirical in vitro validation, we identified 5 key amino acid differences between murine and human ACE2 that mediate SARS-CoV-2 infection, generating a chimeric humanized murine ACE2. Additionally, we examined the ability of the humanized murine ACE2 receptor to permit infection by an additional preemergent group 2B coronavirus, WIV-1, providing evidence for the potential pan-virus capabilities of this chimeric receptor. Finally, we predicted the ability of these determinants to inform host range identification of preemergent coronaviruses by evaluating hot spot contacts between SARS-CoV-2 and additional potential host receptors. Our results identify residue determinants that mediate coronavirus receptor usage and host range for application in SARS-CoV-2 and emerging coronavirus animal model development.IMPORTANCE SARS-CoV-2 (the causative agent of COVID-19) is a major public health threat and one of two related coronaviruses that have caused epidemics in modern history. A method of screening potential infectible hosts for preemergent and future emergent coronaviruses would aid in mounting rapid response and intervention strategies during future emergence events. Here, we evaluated determinants of SARS-CoV-2 receptor interactions, identifying key changes that enable or prevent infection. The analysis detailed in this study will aid in the development of model systems to screen emergent coronaviruses as well as treatments to counteract infections.
Keywords: COVID-19; SARS-CoV-2; coronavirus; host range; receptors; virus-host interactions.
Copyright © 2021 Adams et al.
Figures

References
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Menachery VD, Yount BL, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113:3048–3053. doi: 10.1073/pnas.1517719113. - DOI - PMC - PubMed
-
- Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, Saindane M, Kolykhalov AA, Painter G, Harcourt J, Tamin A, Thornburg NJ, Swanstrom R, Denison MR, Baric RS. 2020. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12:eabb5883. doi: 10.1126/scitranslmed.abb5883. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

