Crystal structure of a far-red-sensing cyanobacteriochrome reveals an atypical bilin conformation and spectral tuning mechanism
- PMID: 33727422
- PMCID: PMC8000052
- DOI: 10.1073/pnas.2025094118
Crystal structure of a far-red-sensing cyanobacteriochrome reveals an atypical bilin conformation and spectral tuning mechanism
Abstract
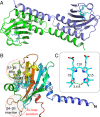
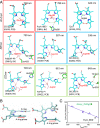
Cyanobacteriochromes (CBCRs) are small, linear tetrapyrrole (bilin)-binding photoreceptors in the phytochrome superfamily that regulate diverse light-mediated adaptive processes in cyanobacteria. More spectrally diverse than canonical red/far-red-sensing phytochromes, CBCRs were thought to be restricted to sensing visible and near UV light until recently when several subfamilies with far-red-sensing representatives (frCBCRs) were discovered. Two of these frCBCRs subfamilies have been shown to incorporate bilin precursors with larger pi-conjugated chromophores, while the third frCBCR subfamily uses the same phycocyanobilin precursor found in the bulk of the known CBCRs. To elucidate the molecular basis of far-red light perception by this third frCBCR subfamily, we determined the crystal structure of the far-red-absorbing dark state of one such frCBCR Anacy_2551g3 from Anabaena cylindrica PCC 7122 which exhibits a reversible far-red/orange photocycle. Determined by room temperature serial crystallography and cryocrystallography, the refined 2.7-Å structure reveals an unusual all-Z,syn configuration of the phycocyanobilin (PCB) chromophore that is considerably less extended than those of previously characterized red-light sensors in the phytochrome superfamily. Based on structural and spectroscopic comparisons with other bilin-binding proteins together with site-directed mutagenesis data, our studies reveal protein-chromophore interactions that are critical for the atypical bathochromic shift. Based on these analyses, we propose that far-red absorption in Anacy_2551g3 is the result of the additive effect of two distinct red-shift mechanisms involving cationic bilin lactim tautomers stabilized by a constrained all-Z,syn conformation and specific interactions with a highly conserved anionic residue.
Keywords: bilin tautomers; bilin-based photoreceptor; far-red-light sensing; optogenetics; serial crystallography.
Conflict of interest statement
The authors declare no competing interest.
Figures


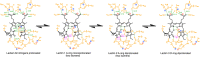
Similar articles
-
Cyanobacteriochromes: A Rainbow of Photoreceptors.Annu Rev Microbiol. 2024 Nov;78(1):61-81. doi: 10.1146/annurev-micro-041522-094613. Epub 2024 Nov 7. Annu Rev Microbiol. 2024. PMID: 38848579 Free PMC article. Review.
-
Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.Proc Natl Acad Sci U S A. 2021 May 18;118(20):e2024583118. doi: 10.1073/pnas.2024583118. Proc Natl Acad Sci U S A. 2021. PMID: 33972439 Free PMC article.
-
A far-red cyanobacteriochrome lineage specific for verdins.Proc Natl Acad Sci U S A. 2020 Nov 10;117(45):27962-27970. doi: 10.1073/pnas.2016047117. Epub 2020 Oct 26. Proc Natl Acad Sci U S A. 2020. PMID: 33106421 Free PMC article.
-
Probing Bilin-Protein Interaction in the Protochromic Photocycle of Cyanobacteriochrome RcaE by Site-Directed Mutagenesis.Plant Cell Physiol. 2025 Feb 28;66(2):181-192. doi: 10.1093/pcp/pcae085. Plant Cell Physiol. 2025. PMID: 39092561
-
Phytochromes and Cyanobacteriochromes: Photoreceptor Molecules Incorporating a Linear Tetrapyrrole Chromophore.Adv Exp Med Biol. 2021;1293:167-187. doi: 10.1007/978-981-15-8763-4_10. Adv Exp Med Biol. 2021. PMID: 33398813 Review.
Cited by
-
Light- and pH-dependent structural changes in cyanobacteriochrome AnPixJg2.Photochem Photobiol Sci. 2022 Apr;21(4):447-469. doi: 10.1007/s43630-022-00204-4. Epub 2022 Apr 8. Photochem Photobiol Sci. 2022. PMID: 35394641
-
Cyanobacteriochromes: A Rainbow of Photoreceptors.Annu Rev Microbiol. 2024 Nov;78(1):61-81. doi: 10.1146/annurev-micro-041522-094613. Epub 2024 Nov 7. Annu Rev Microbiol. 2024. PMID: 38848579 Free PMC article. Review.
-
Red Light Optogenetics in Neuroscience.Front Cell Neurosci. 2022 Jan 3;15:778900. doi: 10.3389/fncel.2021.778900. eCollection 2021. Front Cell Neurosci. 2022. PMID: 35046775 Free PMC article. Review.
-
Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.Proc Natl Acad Sci U S A. 2021 May 18;118(20):e2024583118. doi: 10.1073/pnas.2024583118. Proc Natl Acad Sci U S A. 2021. PMID: 33972439 Free PMC article.
-
Molecular origins of absorption wavelength variation among phycocyanobilin-binding proteins.Biophys J. 2024 Oct 1;123(19):3375-3385. doi: 10.1016/j.bpj.2024.08.001. Epub 2024 Aug 8. Biophys J. 2024. PMID: 39113359 Free PMC article.
References
-
- Ikeuchi M., Ishizuka T., Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7, 1159–1167 (2008). - PubMed
-
- Gan F., et al. ., Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317 (2014). - PubMed
-
- Fushimi K., Narikawa R., Cyanobacteriochromes: Photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 57, 39–46 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

