Deep learning classification of lung cancer histology using CT images
- PMID: 33727623
- PMCID: PMC7943565
- DOI: 10.1038/s41598-021-84630-x
Deep learning classification of lung cancer histology using CT images
Abstract
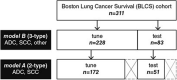
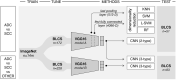
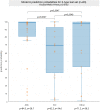
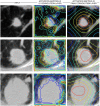
Tumor histology is an important predictor of therapeutic response and outcomes in lung cancer. Tissue sampling for pathologist review is the most reliable method for histology classification, however, recent advances in deep learning for medical image analysis allude to the utility of radiologic data in further describing disease characteristics and for risk stratification. In this study, we propose a radiomics approach to predicting non-small cell lung cancer (NSCLC) tumor histology from non-invasive standard-of-care computed tomography (CT) data. We trained and validated convolutional neural networks (CNNs) on a dataset comprising 311 early-stage NSCLC patients receiving surgical treatment at Massachusetts General Hospital (MGH), with a focus on the two most common histological types: adenocarcinoma (ADC) and Squamous Cell Carcinoma (SCC). The CNNs were able to predict tumor histology with an AUC of 0.71(p = 0.018). We also found that using machine learning classifiers such as k-nearest neighbors (kNN) and support vector machine (SVM) on CNN-derived quantitative radiomics features yielded comparable discriminative performance, with AUC of up to 0.71 (p = 0.017). Our best performing CNN functioned as a robust probabilistic classifier in heterogeneous test sets, with qualitatively interpretable visual explanations to its predictions. Deep learning based radiomics can identify histological phenotypes in lung cancer. It has the potential to augment existing approaches and serve as a corrective aid for diagnosticians.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

