Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations
- PMID: 33728680
- PMCID: PMC8250610
- DOI: 10.1002/1873-3468.14076
Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations
Abstract
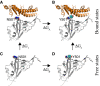
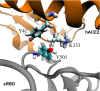
Recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants (B.1.1.7 and B.1351) have emerged harbouring mutations that make them highly contagious. The N501Y mutation within the receptor-binding domain (RBD) of the spike protein of these SARS-CoV-2 variants may enhance binding to the human angiotensin-converting enzyme 2 (hACE2). However, no molecular explanation for such an enhanced affinity has so far been provided. Here, using all-atom molecular dynamics simulations, we show that Y501 in the mutated RBD can be well-coordinated by Y41 and K353 in hACE2 through hydrophobic interactions, which may increase the overall binding affinity of the RBD for hACE2 by approximately 0.81 kcal·mol-1 . The binding dynamics revealed in our study may provide a working model to facilitate the design of more effective antibodies.
Keywords: ACE2; N501Y; SARS-CoV-2; antibody; spike protein.
© 2021 Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

