Mechanisms of repeat-associated non-AUG translation in neurological microsatellite expansion disorders
- PMID: 33729487
- PMCID: PMC8106499
- DOI: 10.1042/BST20200690
Mechanisms of repeat-associated non-AUG translation in neurological microsatellite expansion disorders
Abstract
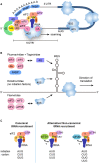
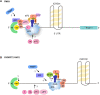
Repeat-associated non-AUG (RAN) translation was discovered in 2011 in spinocerebellar ataxia type 8 (SCA8) and myotonic dystrophy type 1 (DM1). This non-canonical form of translation occurs in all reading frames from both coding and non-coding regions of sense and antisense transcripts carrying expansions of trinucleotide to hexanucleotide repeat sequences. RAN translation has since been reported in 7 of the 53 known microsatellite expansion disorders which mainly present with neurodegenerative features. RAN translation leads to the biosynthesis of low-complexity polymeric repeat proteins with aggregating and cytotoxic properties. However, the molecular mechanisms and protein factors involved in assembling functional ribosomes in absence of canonical AUG start codons remain poorly characterised while secondary repeat RNA structures play key roles in initiating RAN translation. Here, we briefly review the repeat expansion disorders, their complex pathogenesis and the mechanisms of physiological translation initiation together with the known factors involved in RAN translation. Finally, we discuss research challenges surrounding the understanding of pathogenesis and future directions that may provide opportunities for the development of novel therapeutic approaches for this group of incurable neurodegenerative diseases.
Keywords: RAN translation; microsatellite repeat expansion disorders; pathophysiology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
Repeat-Associated Non-ATG Translation in Neurological Diseases.Cold Spring Harb Perspect Biol. 2018 Dec 3;10(12):a033019. doi: 10.1101/cshperspect.a033019. Cold Spring Harb Perspect Biol. 2018. PMID: 29891563 Free PMC article. Review.
-
Repeat-associated non-ATG (RAN) translation in neurological disease.Hum Mol Genet. 2013 Oct 15;22(R1):R45-51. doi: 10.1093/hmg/ddt371. Epub 2013 Aug 4. Hum Mol Genet. 2013. PMID: 23918658 Free PMC article. Review.
-
CAG repeat expansions create splicing acceptor sites and produce aberrant repeat-containing RNAs.Mol Cell. 2024 Feb 15;84(4):702-714.e10. doi: 10.1016/j.molcel.2024.01.006. Epub 2024 Jan 30. Mol Cell. 2024. PMID: 38295802 Free PMC article.
-
Repeat associated non-ATG (RAN) translation: new starts in microsatellite expansion disorders.Curr Opin Genet Dev. 2014 Jun;26:6-15. doi: 10.1016/j.gde.2014.03.002. Epub 2014 May 22. Curr Opin Genet Dev. 2014. PMID: 24852074 Free PMC article. Review.
-
Non-ATG-initiated translation directed by microsatellite expansions.Proc Natl Acad Sci U S A. 2011 Jan 4;108(1):260-5. doi: 10.1073/pnas.1013343108. Epub 2010 Dec 20. Proc Natl Acad Sci U S A. 2011. PMID: 21173221 Free PMC article.
Cited by
-
The human DEAD-box helicase DDX3X as a regulator of mRNA translation.Front Cell Dev Biol. 2022 Oct 25;10:1033684. doi: 10.3389/fcell.2022.1033684. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36393867 Free PMC article. Review.
-
Structural and Dynamical Properties of Nucleic Acid Hairpins Implicated in Trinucleotide Repeat Expansion Diseases.Biomolecules. 2024 Oct 10;14(10):1278. doi: 10.3390/biom14101278. Biomolecules. 2024. PMID: 39456210 Free PMC article. Review.
-
Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications.Nat Rev Genet. 2024 Jul;25(7):476-499. doi: 10.1038/s41576-024-00696-z. Epub 2024 Mar 11. Nat Rev Genet. 2024. PMID: 38467784 Review.
-
Proteinopathies as Hallmarks of Impaired Gene Expression, Proteostasis and Mitochondrial Function in Amyotrophic Lateral Sclerosis.Front Neurosci. 2021 Dec 23;15:783624. doi: 10.3389/fnins.2021.783624. eCollection 2021. Front Neurosci. 2021. PMID: 35002606 Free PMC article. Review.
-
Dissecting the mechanism of NOP56 GGCCUG repeat-associated non-AUG translation using cell-free translation systems.J Biol Chem. 2025 Apr;301(4):108360. doi: 10.1016/j.jbc.2025.108360. Epub 2025 Feb 25. J Biol Chem. 2025. PMID: 40015643 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

