VLP-factory™ and ADDomer© : Self-assembling Virus-Like Particle (VLP) Technologies for Multiple Protein and Peptide Epitope Display
- PMID: 33729713
- PMCID: PMC9733710
- DOI: 10.1002/cpz1.55
VLP-factory™ and ADDomer© : Self-assembling Virus-Like Particle (VLP) Technologies for Multiple Protein and Peptide Epitope Display
Erratum in
-
Group Correction Statement (Data Availability Statements).Curr Protoc. 2022 Aug;2(8):e552. doi: 10.1002/cpz1.552. Curr Protoc. 2022. PMID: 36005902 Free PMC article. No abstract available.
-
Group Correction Statement (Conflict of Interest Statements).Curr Protoc. 2022 Aug;2(8):e551. doi: 10.1002/cpz1.551. Curr Protoc. 2022. PMID: 36005903 Free PMC article. No abstract available.
Abstract
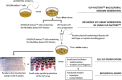
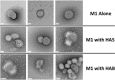
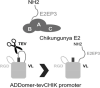
Virus-like particles (VLPs) play a prominent role in vaccination as safe and highly versatile alternatives to attenuated or inactivated viruses or subunit vaccines. We present here two innovations, VLP-factory™ and ADDomer© , for creating VLPs displaying entire proteins or peptide epitopes as antigens, respectively, to enable efficient vaccination. For producing these VLPs, we use MultiBac, a baculovirus expression vector system (BEVS) that we developed for producing complex protein biologics in insect cells transfected with an engineered baculovirus. VLPs are protein assemblies that share features with viruses but are devoid of genetic material, and thus considered safe. VLP-factory™ represents a customized MultiBac baculovirus tailored to produce enveloped VLPs based on the M1 capsid protein of influenza virus. We apply VLP-factory™ to create an array of influenza-derived VLPs presenting functional mutant influenza hemagglutinin (HA) glycoprotein variants. Moreover, we describe MultiBac-based production of ADDomer© , a synthetic self-assembling adenovirus-derived protein-based VLP platform designed to display multiple copies of pathogenic epitopes at the same time on one particle for highly efficient vaccination. © 2021 The Authors. Basic Protocol 1: VLP-factory™ baculoviral genome generation Basic Protocol 2: Influenza VLP array generation using VLP-factory™ Basic Protocol 3: Influenza VLP purification Basic Protocol 4: ADDomer© BioBrick design, expression, and purification Basic Protocol 5: ADDomer© candidate vaccines against infectious diseases.
Keywords: MultiBac; antigenic epitope; baculovirus expression vector system (BEVS); immunization; protein and peptide display; vaccine; virus-like particle (VLP).
© 2021 The Authors.
Conflict of interest statement
I.B., F.G. and D.F. declare conflict or interest related to this correspondence. I.B. and F.G. are shareholders of Imophoron Ltd, Imophoron owns patents and trademarks related to ADDomer and its applications. I.B. and D.F. are shareholders of Geneva Biotech SARL. Geneva Biotech owns patents related to the MultiBac system and its applications including VLP‐Factory.
Figures






Similar articles
-
Virus-like particle vaccines with epitopes from porcine epidemic virus and transmissible gastroenteritis virus incorporated into self-assembling ADDomer platform provide clinical immune responses in piglets.Front Immunol. 2023 Oct 24;14:1251001. doi: 10.3389/fimmu.2023.1251001. eCollection 2023. Front Immunol. 2023. PMID: 37942329 Free PMC article.
-
High-Throughput Production of Influenza Virus-Like Particle (VLP) Array by Using VLP-factory™, a MultiBac Baculoviral Genome Customized for Enveloped VLP Expression.Methods Mol Biol. 2019;2025:213-226. doi: 10.1007/978-1-4939-9624-7_10. Methods Mol Biol. 2019. PMID: 31267455
-
Structural basis for the development of avian virus capsids that display influenza virus proteins and induce protective immunity.J Virol. 2015 Mar;89(5):2563-74. doi: 10.1128/JVI.03025-14. Epub 2014 Dec 17. J Virol. 2015. PMID: 25520499 Free PMC article.
-
Development of virus-like particle technology from small highly symmetric to large complex virus-like particle structures.Intervirology. 2013;56(3):141-65. doi: 10.1159/000346773. Epub 2013 Apr 16. Intervirology. 2013. PMID: 23594863 Review.
-
Virus-like particle (VLP)-based vaccines for pandemic influenza: performance of a VLP vaccine during the 2009 influenza pandemic.Hum Vaccin Immunother. 2012 Mar;8(3):411-4. doi: 10.4161/hv.18757. Epub 2012 Feb 14. Hum Vaccin Immunother. 2012. PMID: 22330956 Free PMC article. Review.
Cited by
-
In vitro generated antibodies guide thermostable ADDomer nanoparticle design for nasal vaccination and passive immunization against SARS-CoV-2.Antib Ther. 2023 Oct 17;6(4):277-297. doi: 10.1093/abt/tbad024. eCollection 2023 Oct. Antib Ther. 2023. PMID: 38075238 Free PMC article.
-
Virus-like particle vaccines with epitopes from porcine epidemic virus and transmissible gastroenteritis virus incorporated into self-assembling ADDomer platform provide clinical immune responses in piglets.Front Immunol. 2023 Oct 24;14:1251001. doi: 10.3389/fimmu.2023.1251001. eCollection 2023. Front Immunol. 2023. PMID: 37942329 Free PMC article.
-
Development of virus-like particles-based vaccines against coronaviruses.Biotechnol Prog. 2022 Nov;38(6):e3292. doi: 10.1002/btpr.3292. Epub 2022 Aug 19. Biotechnol Prog. 2022. PMID: 35932092 Free PMC article. Review.
-
Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.Structure. 2024 Mar 7;32(3):342-351.e6. doi: 10.1016/j.str.2023.12.010. Epub 2024 Jan 9. Structure. 2024. PMID: 38198950 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

