Structural analysis of viral ExoN domains reveals polyphyletic hijacking events
- PMID: 33730017
- PMCID: PMC7968707
- DOI: 10.1371/journal.pone.0246981
Structural analysis of viral ExoN domains reveals polyphyletic hijacking events
Abstract
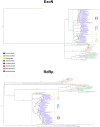
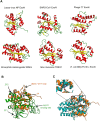
Nidoviruses and arenaviruses are the only known RNA viruses encoding a 3'-5' exonuclease domain (ExoN). The proofreading activity of the ExoN domain has played a key role in the growth of nidoviral genomes, while in arenaviruses this domain partakes in the suppression of the host innate immune signaling. Sequence and structural homology analyses suggest that these proteins have been hijacked from cellular hosts many times. Analysis of the available nidoviral ExoN sequences reveals a high conservation level comparable to that of the viral RNA-dependent RNA polymerases (RdRp), which are the most conserved viral proteins. Two highly preserved zinc fingers are present in all nidoviral exonucleases, while in the arenaviral protein only one zinc finger can be identified. This is in sharp contrast with the reported lack of zinc fingers in cellular ExoNs, and opens the possibility of therapeutic strategies in the struggle against COVID-19.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- World Health Organization (WHO). 2021. www.who.int
-
- Cheng VCC, Lau SKP, Woo PCY, Yuen KY. Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection. Clin Microbiol Rev. 2007. October;20(4):660–94. Available from: https://cmr.asm.org/content/20/4/660 - PMC - PubMed
-
- Enjuanes L, Gorbalenya AE, de Groot RJ, Cowley JA, Ziebuhr J, Snijder EJ. Nidovirales. In: Encyclopedia of Virology. Elsevier; 2008. p. 419–30. https://linkinghub.elsevier.com/retrieve/pii/B9780123744104007755
-
- Hoet AE, Horzinek MC. Torovirus. In: Encyclopedia of Virology. Elsevier; 2008. p. 151–7. https://linkinghub.elsevier.com/retrieve/pii/B9780123744104005161
-
- Walker PJ, Sittidilokratna N. Yellow Head Virus. In: Encyclopedia of Virology. Elsevier; 2008. p. 476–83. https://linkinghub.elsevier.com/retrieve/pii/B9780123744104007792
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

