Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes
- PMID: 33730024
- PMCID: PMC8007021
- DOI: 10.1371/journal.pbio.3001143
Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes
Abstract
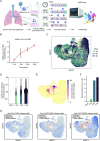
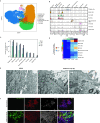
There are currently limited Food and Drug Administration (FDA)-approved drugs and vaccines for the treatment or prevention of Coronavirus Disease 2019 (COVID-19). Enhanced understanding of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection and pathogenesis is critical for the development of therapeutics. To provide insight into viral replication, cell tropism, and host-viral interactions of SARS-CoV-2, we performed single-cell (sc) RNA sequencing (RNA-seq) of experimentally infected human bronchial epithelial cells (HBECs) in air-liquid interface (ALI) cultures over a time course. This revealed novel polyadenylated viral transcripts and highlighted ciliated cells as a major target at the onset of infection, which we confirmed by electron and immunofluorescence microscopy. Over the course of infection, the cell tropism of SARS-CoV-2 expands to other epithelial cell types including basal and club cells. Infection induces cell-intrinsic expression of type I and type III interferons (IFNs) and interleukin (IL)-6 but not IL-1. This results in expression of interferon-stimulated genes (ISGs) in both infected and bystander cells. This provides a detailed characterization of genes, cell types, and cell state changes associated with SARS-CoV-2 infection in the human airway.
Conflict of interest statement
Yale University (CBW) has a patent pending related to this work entitled: “Compounds and Compositions for Treating, Ameliorating, and/or Preventing SARS-CoV-2 Infection and/or Complications Thereof.” Yale University has committed to rapidly executable non-exclusive royalty-free licenses to intellectual property rights for the purpose of making and distributing products to prevent, diagnose and treat COVID-19 infection during the pandemic and for a short period thereafter.
Figures



Update of
-
Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium.bioRxiv [Preprint]. 2020 Jul 13:2020.05.06.081695. doi: 10.1101/2020.05.06.081695. bioRxiv. 2020. Update in: PLoS Biol. 2021 Mar 17;19(3):e3001143. doi: 10.1371/journal.pbio.3001143. PMID: 32511382 Free PMC article. Updated. Preprint.
Comment in
-
The first few days of a SARS-CoV-2 infection viewed at single-cell resolution.PLoS Biol. 2021 Apr 26;19(4):e3001217. doi: 10.1371/journal.pbio.3001217. eCollection 2021 Apr. PLoS Biol. 2021. PMID: 33901166 Free PMC article.
References
-
- World Health Organization. Novel Coronavirus (2019-nCoV): situation report, 22. 2020 [cited 4 Sep 2020]. Available from: https://apps.who.int/iris/handle/10665/330991
Publication types
MeSH terms
Grants and funding
- T32 AI055403/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- UL1 TR001863/TR/NCATS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R21 AI133440/AI/NIAID NIH HHS/United States
- K08 AI119139/AI/NIAID NIH HHS/United States
- R01 HL138540/HL/NHLBI NIH HHS/United States
- K08 AI128043/AI/NIAID NIH HHS/United States
- K08 HL136898/HL/NHLBI NIH HHS/United States
- F30 CA250249/CA/NCI NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- R01 AI141609/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

