Advances and Opportunities in Single-Cell Transcriptomics for Plant Research
- PMID: 33730513
- PMCID: PMC7611048
- DOI: 10.1146/annurev-arplant-081720-010120
Advances and Opportunities in Single-Cell Transcriptomics for Plant Research
Abstract
Single-cell approaches are quickly changing our view on biological systems by increasing the spatiotemporal resolution of our analyses to the level of the individual cell. The field of plant biology has fully embraced single-cell transcriptomics and is rapidly expanding the portfolio of available technologies and applications. In this review, we give an overview of the main advances in plant single-cell transcriptomics over the past few years and provide the reader with an accessible guideline covering all steps, from sample preparation to data analysis. We end by offering a glimpse of how these technologies will shape and accelerate plant-specific research in the near future.
Keywords: cell atlas; cell trajectory; plant transcriptomics; protoplast; scRNA-seq.
Conflict of interest statement
Figures
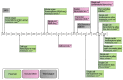

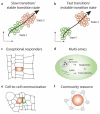
References
-
- Bailey-Serres J. Microgenomics: Genome-Scale, Cell-Specific Monitoring of Multiple Gene Regulation Tiers. Annu Rev Plant Biol. 2013;64(1):293–325. - PubMed
-
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37(1):38–44. - PubMed
-
- et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

