Antigen presentation between T cells drives Th17 polarization under conditions of limiting antigen
- PMID: 33730591
- PMCID: PMC7972993
- DOI: 10.1016/j.celrep.2021.108861
Antigen presentation between T cells drives Th17 polarization under conditions of limiting antigen
Abstract
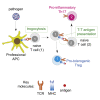
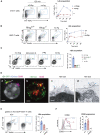
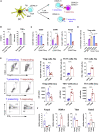
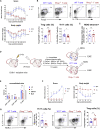
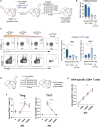
T cells form immunological synapses with professional antigen-presenting cells (APCs) resulting in T cell activation and the acquisition of peptide antigen-MHC (pMHC) complexes from the plasma membrane of the APC. They thus become APCs themselves. We investigate the functional outcome of T-T cell antigen presentation by CD4 T cells and find that the antigen-presenting T cells (Tpres) predominantly differentiate into regulatory T cells (Treg), whereas T cells that have been stimulated by Tpres cells predominantly differentiate into Th17 pro-inflammatory cells. Using mice deficient in pMHC uptake by T cells, we show that T-T antigen presentation is important for the development of experimental autoimmune encephalitis and Th17 cell differentiation in vivo. By varying the professional APC:T cell ratio, we can modulate Treg versus Th17 differentiation in vitro and in vivo, suggesting that T-T antigen presentation underlies proinflammatory responses in conditions of antigen scarcity.
Keywords: Th17; Treg; antigen presentation by T cells; limiting antigen; trogocytosis; vaccine dosing.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Arnold P.Y., Mannie M.D. Vesicles bearing MHC class II molecules mediate transfer of antigen from antigen-presenting cells to CD4+ T cells. Eur. J. Immunol. 1999;29:1363–1373. - PubMed
-
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. - PubMed
-
- Billeskov R., Wang Y., Solaymani-Mohammadi S., Frey B., Kulkarni S., Andersen P., Agger E.M., Sui Y., Berzofsky J.A. Low Antigen Dose in Adjuvant-Based Vaccination Selectively Induces CD4 T Cells with Enhanced Functional Avidity and Protective Efficacy. J. Immunol. 2017;198:3494–3506. - PMC - PubMed
-
- Borroto A., Reyes-Garau D., Jiménez M.A., Carrasco E., Moreno B., Martínez-Pasamar S., Cortés J.R., Perona A., Abia D., Blanco S. First-in-class inhibitor of the T cell receptor for the treatment of autoimmune diseases. Sci. Transl. Med. 2016;8:370ra184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

