Structural and functional analysis of tomato sterol C22 desaturase
- PMID: 33731007
- PMCID: PMC7972189
- DOI: 10.1186/s12870-021-02898-7
Structural and functional analysis of tomato sterol C22 desaturase
Abstract
Background: Sterols are structural and functional components of eukaryotic cell membranes. Plants produce a complex mixture of sterols, among which β-sitosterol, stigmasterol, campesterol, and cholesterol in some Solanaceae, are the most abundant species. Many reports have shown that the stigmasterol to β-sitosterol ratio changes during plant development and in response to stresses, suggesting that it may play a role in the regulation of these processes. In tomato (Solanum lycopersicum), changes in the stigmasterol to β-sitosterol ratio correlate with the induction of the only gene encoding sterol C22-desaturase (C22DES), the enzyme specifically involved in the conversion of β-sitosterol to stigmasterol. However, despite the biological interest of this enzyme, there is still a lack of knowledge about several relevant aspects related to its structure and function.
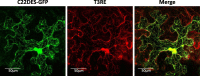
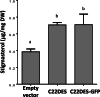
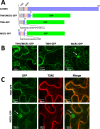
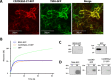
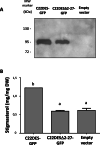
Results: In this study we report the subcellular localization of tomato C22DES in the endoplasmic reticulum (ER) based on confocal fluorescence microscopy and cell fractionation analyses. Modeling studies have also revealed that C22DES consists of two well-differentiated domains: a single N-terminal transmembrane-helix domain (TMH) anchored in the ER-membrane and a globular (or catalytic) domain that is oriented towards the cytosol. Although TMH is sufficient for the targeting and retention of the enzyme in the ER, the globular domain may also interact and be retained in the ER in the absence of the N-terminal transmembrane domain. The observation that a truncated version of C22DES lacking the TMH is enzymatically inactive revealed that the N-terminal membrane domain is essential for enzyme activity. The in silico analysis of the TMH region of plant C22DES revealed several structural features that could be involved in substrate recognition and binding.
Conclusions: Overall, this study contributes to expand the current knowledge on the structure and function of plant C22DES and to unveil novel aspects related to plant sterol metabolism.
Keywords: Cytochrome P450; Sterol C22-desaturase, endoplasmic reticulum; Sterol metabolism, Stigmasterol; Tomato; β-Sitosterol.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Demel RA, De Kruyff B. The function of sterols in membranes. Biochem Biophys Acta. 1976;457:109–132. - PubMed
-
- Hartmann-Bouillon M-A, Benveniste P. Sterol biosynthetic capability of purified membrane fractions from maize coleoptiles. Phytochemistry. 1978;17:1037–1042. doi: 10.1016/S0031-9422(00)94275-4. - DOI
-
- Hartmann M-A. Plant sterols and the membrane environment. Trends Plant Sci. 1998;3(5):170–175. doi: 10.1016/S1360-1385(98)01233-3. - DOI

