CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated β-catenin stabilization
- PMID: 33731207
- PMCID: PMC7972245
- DOI: 10.1186/s13046-021-01896-9
CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated β-catenin stabilization
Abstract
Background: Circular RNAs (circRNAs) have emerged as a new subclass of regulatory RNAs that play critical roles in various cancers. Cancer stem cells (CSCs), a small subset of cancer cells, are believed to possess the capacities to initiate tumorigenesis and promote progression. Although accumulating evidence has suggested that cells with CSC-like properties are crucial for the malignancy of gastric cancer (GC), it remains unclear whether circRNAs are related to the acquisition of CSC-like properties in GC.
Methods: CircFAM73A expression was analyzed by GEO datasets and verified in GC samples. The roles of circFAM73A in GC cell proliferation, migration, cisplatin resistance, and CSC-like properties were determined by a series of functional experiments both in vitro and in vivo. RNA pulldown was used to explore the miRNAs and proteins binding to circFAM73A. Bioinformatic analysis and experimental verification confirmed the downstream targets of circFAM73A. The regulation of circFAM73A by HMGA2 was verified by ChIP and RIP assays.
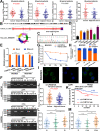
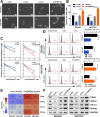
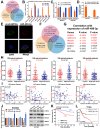
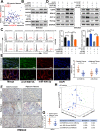
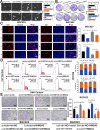
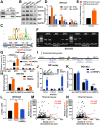
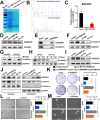
Results: Elevated circFAM73A expression was confirmed in GC tissues, and higher circFAM73A predicted poor prognosis in GC patients. The upregulation of circFAM73A enhanced CSC-like properties in GC, thus facilitating cell proliferation, migration, and cisplatin resistance. Mechanistically, circFAM73A promoted GC malignancy by regulating miR-490-3p/HMGA2 in a positive feedback loop and recruiting HNRNPK to facilitate β-catenin stabilization. Moreover, HMGA2 further enhanced E2F1 and HNRNPL activity, which in turn promoted circFAM73A expression.
Conclusions: Our work demonstrates the crucial role of circFAM73A in the CSC-like properties of GC and uncovers a positive feedback loop in circFAM73A regulation that leads to the progression of gastric cancer, which may provide new insights into circRNA-based diagnostic and therapeutic strategies.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Circ_0000267 promotes gastric cancer progression via sponging MiR-503-5p and regulating HMGA2 expression.Mol Genet Genomic Med. 2020 Feb;8(2):e1093. doi: 10.1002/mgg3.1093. Epub 2019 Dec 17. Mol Genet Genomic Med. 2020. PMID: 31845519 Free PMC article.
-
LncRNA FEZF1-AS1 Modulates Cancer Stem Cell Properties of Human Gastric Cancer Through miR-363-3p/HMGA2.Cell Transplant. 2020 Jan-Dec;29:963689720925059. doi: 10.1177/0963689720925059. Cell Transplant. 2020. Retraction in: Cell Transplant. 2024 Jan-Dec;33:9636897241298459. doi: 10.1177/09636897241298459. PMID: 32638620 Free PMC article. Retracted.
-
Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression.Int J Biol Sci. 2019 Apr 22;15(5):1091-1103. doi: 10.7150/ijbs.31533. eCollection 2019. Int J Biol Sci. 2019. PMID: 31182928 Free PMC article.
-
HMGA2 regulation by miRNAs in cancer: Affecting cancer hallmarks and therapy response.Pharmacol Res. 2023 Apr;190:106732. doi: 10.1016/j.phrs.2023.106732. Epub 2023 Mar 15. Pharmacol Res. 2023. PMID: 36931542 Review.
-
CircRNAs as upstream regulators of miRNA//HMGA2 axis in human cancer.Pharmacol Ther. 2024 Nov;263:108711. doi: 10.1016/j.pharmthera.2024.108711. Epub 2024 Aug 31. Pharmacol Ther. 2024. PMID: 39222752 Review.
Cited by
-
Emerging roles of circular RNAs in stem cells.Genes Dis. 2022 May 31;10(5):1920-1936. doi: 10.1016/j.gendis.2022.05.015. eCollection 2023 Sep. Genes Dis. 2022. PMID: 37492713 Free PMC article. Review.
-
The multifaceted roles of circular RNAs in cancer hallmarks: From mechanisms to clinical implications.Mol Ther Nucleic Acids. 2024 Jul 20;35(3):102286. doi: 10.1016/j.omtn.2024.102286. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39188305 Free PMC article. Review.
-
β-catenin as a key regulator of the cisplatin response in tumor cells.Clin Exp Med. 2025 Jun 15;25(1):206. doi: 10.1007/s10238-025-01757-1. Clin Exp Med. 2025. PMID: 40517336 Free PMC article. Review.
-
Natriuretic peptide receptor a promotes gastric malignancy through angiogenesis process.Cell Death Dis. 2021 Oct 20;12(11):968. doi: 10.1038/s41419-021-04266-7. Cell Death Dis. 2021. PMID: 34671022 Free PMC article.
-
Deciphering the impact of circRNA-mediated autophagy on tumor therapeutic resistance: a novel perspective.Cell Mol Biol Lett. 2024 Apr 26;29(1):60. doi: 10.1186/s11658-024-00571-z. Cell Mol Biol Lett. 2024. PMID: 38671354 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

