GREB1: An evolutionarily conserved protein with a glycosyltransferase domain links ERα glycosylation and stability to cancer
- PMID: 33731348
- PMCID: PMC7968844
- DOI: 10.1126/sciadv.abe2470
GREB1: An evolutionarily conserved protein with a glycosyltransferase domain links ERα glycosylation and stability to cancer
Abstract
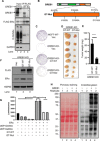
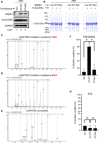
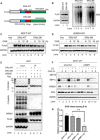
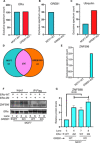
What covalent modifications control the temporal ubiquitination of ERα and hence the duration of its transcriptional activity remain poorly understood. We show that GREB1, an ERα-inducible enzyme, catalyzes O-GlcNAcylation of ERα at residues T553/S554, which stabilizes ERα protein by inhibiting association with the ubiquitin ligase ZNF598. Loss of GREB1-mediated glycosylation of ERα results in reduced cellular ERα levels and insensitivity to estrogen. Higher GREB1 expression in ERα+ve breast cancer is associated with greater survival in response to tamoxifen, an ERα agonist. Mice lacking Greb1 exhibit growth and fertility defects reminiscent of phenotypes in ERα-null mice. In summary, this study identifies GREB1, a protein with an evolutionarily conserved domain related to DNA-modifying glycosyltransferases of bacteriophages and kinetoplastids, as the first inducible and the only other (apart from OGT) O-GlcNAc glycosyltransferase in mammalian cytoplasm and ERα as its first substrate.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- Hewitt S. C., Korach K. S., Oestrogen receptor knockout mice: Roles for oestrogen receptors alpha and beta in reproductive tissues. Reproduction 125, 143–149 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

