Striatal Direct Pathway Targets Npas1+ Pallidal Neurons
- PMID: 33731445
- PMCID: PMC8176753
- DOI: 10.1523/JNEUROSCI.2306-20.2021
Striatal Direct Pathway Targets Npas1+ Pallidal Neurons
Abstract
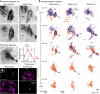
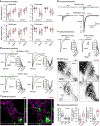
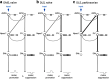
The classic basal ganglia circuit model asserts a complete segregation of the two striatal output pathways. Empirical data argue that, in addition to indirect-pathway striatal projection neurons (iSPNs), direct-pathway striatal projection neurons (dSPNs) innervate the external globus pallidus (GPe). However, the functions of the latter were not known. In this study, we interrogated the organization principles of striatopallidal projections and their roles in full-body movement in mice (both males and females). In contrast to the canonical motor-promoting response of dSPNs in the dorsomedial striatum (DMSdSPNs), optogenetic stimulation of dSPNs in the dorsolateral striatum (DLSdSPNs) suppressed locomotion. Circuit analyses revealed that dSPNs selectively target Npas1+ neurons in the GPe. In a chronic 6-hydroxydopamine lesion model of Parkinson's disease, the dSPN-Npas1+ projection was dramatically strengthened. As DLSdSPN-Npas1+ projection suppresses movement, the enhancement of this projection represents a circuit mechanism for the hypokinetic symptoms of Parkinson's disease that has not been previously considered. In sum, our results suggest that dSPN input to the GPe is a critical circuit component that is involved in the regulation of movement in both healthy and parkinsonian states.SIGNIFICANCE STATEMENT In the classic basal ganglia model, the striatum is described as a divergent structure: it controls motor and adaptive functions through two segregated, opposing output streams. However, the experimental results that show the projection from direct-pathway neurons to the external pallidum have been largely ignored. Here, we showed that this striatopallidal subpathway targets a select subset of neurons in the external pallidum and is motor-suppressing. We found that this subpathway undergoes changes in a Parkinson's disease model. In particular, our results suggest that the increase in strength of this subpathway contributes to the slowness or reduced movements observed in Parkinson's disease.
Keywords: 6-OHDA; GABAergic inhibition; arkypallidal neurons; body kinematics; hypokinesia; movement dynamics.
Copyright © 2021 the authors.
Figures
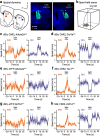

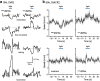
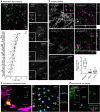
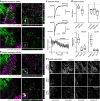

References
-
- Abdi A, Mallet N, Mohamed FY, Sharott A, Dodson PD, Nakamura KC, Suri S, Avery SV, Larvin JT, Garas FN, Garas SN, Vinciati F, Morin S, Bezard E, Baufreton J, Magill PJ (2015) Prototypic and arkypallidal neurons in the dopamine-intact external globus pallidus. J Neurosci 35:6667–6688. 10.1523/JNEUROSCI.4662-14.2015 - DOI - PMC - PubMed
-
- Abecassis ZA, Berceau BL, Win PH, Garcia D, Xenias HS, Cui Q, Pamukcu A, Cherian S, Hernandez VM, Chon U, Lim BK, Kim Y, Justice NJ, Awatramani R, Hooks BM, Gerfen CR, Boca SM, Chan CS (2020) Npas1(+)-Nkx2.1(+) neurons are an integral part of the cortico-pallido-cortical loop. J Neurosci 40:743–768. 10.1523/JNEUROSCI.1199-19.2019 - DOI - PMC - PubMed
-
- Albin RL, Young AB, Penney JB (1995) The functional anatomy of disorders of the basal ganglia. Trends Neurosci 18:63–64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
